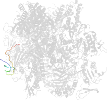
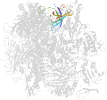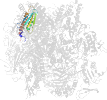Lineage for d2vumj1 (2vum J:1-65)
- Root: SCOPe 2.08

Class a: All alpha proteins [46456] (290 folds) 
Fold a.4: DNA/RNA-binding 3-helical bundle [46688] (14 superfamilies)
core: 3-helices; bundle, closed or partly opened, right-handed twist; up-and down
Superfamily a.4.11: RNA polymerase subunit RPB10 [46924] (1 family) 
automatically mapped to Pfam PF01194
Family a.4.11.1: RNA polymerase subunit RPB10 [46925] (2 proteins)
Zn-binding site is near the N-terminus
Protein RNA polymerase subunit RPB10 [46926] (3 species) 
Species Baker's yeast (Saccharomyces cerevisiae) [TaxId:4932] [63490] (29 PDB entries)
Uniprot P22139; part of multichain biological unit
Domain d2vumj1: 2vum J:1-65 [153554]
Other proteins in same PDB: d2vuma1, d2vumb1, d2vumd1, d2vumf1, d2vumg1, d2vumh1, d2vumk1, d2vuml1
automatically matched to d1i3qj_
protein/DNA complex; protein/RNA complex; complexed with mg, zn
Details for d2vumj1
PDB Entry: 2vum (more details), 3.4 Å
PDB Description: alpha-amanitin inhibited complete rna polymerase ii elongation complex
PDB Compounds: (J:) DNA-directed RNA polymerases I, II, and III subunit RPABC5SCOPe Domain Sequences for d2vumj1:
Sequence; same for both SEQRES and ATOM records: (download)
>d2vumj1 a.4.11.1 (J:1-65) RNA polymerase subunit RPB10 {Baker's yeast (Saccharomyces cerevisiae) [TaxId: 4932]}
mivpvrcfscgkvvgdkwesylnllqedeldegtalsrlglkryccrrmilthvdliekf
lrynp
SCOPe Domain Coordinates for d2vumj1:
Click to download the PDB-style file with coordinates for d2vumj1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d2vumj1:
- d2vumj1 first appeared in SCOP 1.75
- d2vumj1 appears in SCOPe 2.07

 View in 3D
View in 3D