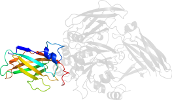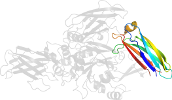Lineage for d1g0da4 (1g0d A:141-461)
- Root: SCOPe 2.01

Class d: Alpha and beta proteins (a+b) [53931] (376 folds) 
Fold d.3: Cysteine proteinases [54000] (1 superfamily)
consists of one alpha-helix and 4 strands of antiparallel beta-sheet and contains the catalytic triad Cys-His-Asn
Superfamily d.3.1: Cysteine proteinases [54001] (23 families) 
the constitute families differ by insertion into and circular permutation of the common catalytic core made of one alpha-helix and 3-strands of beta-sheet
Family d.3.1.4: Transglutaminase core [54044] (3 proteins) 
Protein Transglutaminase catalytic domain [54045] (4 species) 
Species Red sea bream (Chrysophrys major) [TaxId:143350] [64204] (1 PDB entry) 
Domain d1g0da4: 1g0d A:141-461 [60169]
Other proteins in same PDB: d1g0da1, d1g0da2, d1g0da3
complexed with so4
Details for d1g0da4
PDB Entry: 1g0d (more details), 2.5 Å
PDB Description: crystal structure of red sea bream transglutaminase
PDB Compounds: (A:) protein-glutamine gamma-glutamyltransferaseSCOPe Domain Sequences for d1g0da4:
Sequence; same for both SEQRES and ATOM records: (download)
>d1g0da4 d.3.1.4 (A:141-461) Transglutaminase catalytic domain {Red sea bream (Chrysophrys major) [TaxId: 143350]}
dmvylpdesklqeyvmnedgviymgtwdyirsipwnygqfedyvmdicfevldnspaalk
nsemdiehrsdpvyvgrtitamvnsngdrgvltgrweepytdgvapyrwtgsvpilqqws
kagvrpvkygqcwvfaavactvlrclgiptrpitnfasahdvdgnlsvdfllnerlesld
srqrsdsswnfhcwveswmsredlpegndgwqvldptpqelsdgefccgpcpvaaikegn
lgvkydapfvfaevnadtiywivqkdgqrrkitedhasvgknistksvygnhredvtlhy
kypegsqkerevykkagrrvt
SCOPe Domain Coordinates for d1g0da4:
Click to download the PDB-style file with coordinates for d1g0da4.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1g0da4:
- d1g0da4 first appeared in SCOP 1.57
- d1g0da4 appears in SCOP 1.75
- d1g0da4 appears in SCOPe 2.02
- d1g0da4 appears in the current release, SCOPe 2.08

 View in 3D
View in 3D