SCOPe 2.01 help
Overview
Structural Classification of Proteins — extended (SCOPe) is a database of protein structural relationships that extends the SCOP database. SCOP is a (mostly) manually curated ordering of domains from the majority of proteins of known structure in a hierarchy according to structural and evolutionary relationships. Development of SCOP (version 1) concluded with SCOP 1.75, released in June 2009. SCOPe extends the SCOP database, using a combination of manual curation and rigorously validated automated methods to classify many newer PDB structures. Our goal in implementing automation is to extend SCOPe to include structures that can be classified in the SCOP hierarchy, without sacrificing the reliability of the database that was developed through years of expert curation.
SCOPe also incorporates and updates the Astral compendium. Astral provides several databases and tools to aid in the analysis of the protein structures classified in SCOP, particularly through the use of their sequences. Here is a link to more info about Astral. New releases of Astral are derived from SCOPe.
We have rigorously benchmarked our automated methods to ensure that they are as accurate as manual curation, though there are many proteins to which our methods cannot be applied. SCOPe also corrects some errors in SCOP. SCOPe aims to be backward compatible with SCOP, providing the same parseable files and a history of changes between all stable SCOP and SCOPe releases (available on the Stats & History page).
The SCOPe website provides integrated access to data found in all releases of the SCOP and Astral databases that feature stable identifiers.
The SCOP hierarchy
- Applies to: SCOP version 1.55 through current release
- Reference: 5

For backward compatibility, SCOPe entries are arranged in the same hierarchy as SCOP (version 1) entries.
By analogy with taxonomy, SCOP was created as a hierarchy of several levels where the fundamental unit of classification is a domain in the experimentally determined protein structure. Starting at the bottom, the hierarchy of SCOP domains comprises the following levels:
- Species representing a distinct protein sequence and its naturally occurring or artificially created variants.
- Protein grouping together similar sequences of essentially the same functions that either originate from different biological species or represent different isoforms within the same species
- Family containing proteins with similar sequences but typically distinct functions
- Superfamily bridging together protein families with common functional and structural features inferred to be from a common evolutionary ancestor.
- Folds grouping structurally similar superfamilies.
- Classes based mainly on secondary structure content and organization.
Only the first seven classes are true classes. Because traditionally the aim of SCOP has been to classify every residue in the PDB, the remaining classes were maintained as placeholders to keep track of portions of a PDB structure that were not appropriate to include in SCOP. The SCOPe automated classification methods do not currently add entries to classes other than the first seven.
The multi-domain class is a special class. It contains chains with multiple domains that haven't been observed in a different context.
Changes to SCOP(e) design and size
- Applies to: SCOP version 1.55 through current release
- References: 1-5,8
The figure below shows the number of structures in the PDB and
SCOP(e) at the time of
each SCOP(e) release. Extended
horizontal lines start at the freeze date for
each SCOP(e) release and show the
number of PDB entries available on that date. (The "freeze date"
is the last date for PDB entries to be released and still classified
in a given SCOP(e) release. Prior to
SCOP 1.73, all protein structures available on
the freeze date were manually classified.)

The figure below, adapted from
[3],
illustrates how SCOP(e)
has changed over the years. The length of the vertical line for
each release is proportional to the number of entries classified. The angle
of the blue baseline between releases reflects the degree of
divergence from comprehensive and fully manually curated
releases. The angle is based on the percentage of PDB entries on the
freeze date that were not classified in the release, and the extent
to which entries were manually classified.
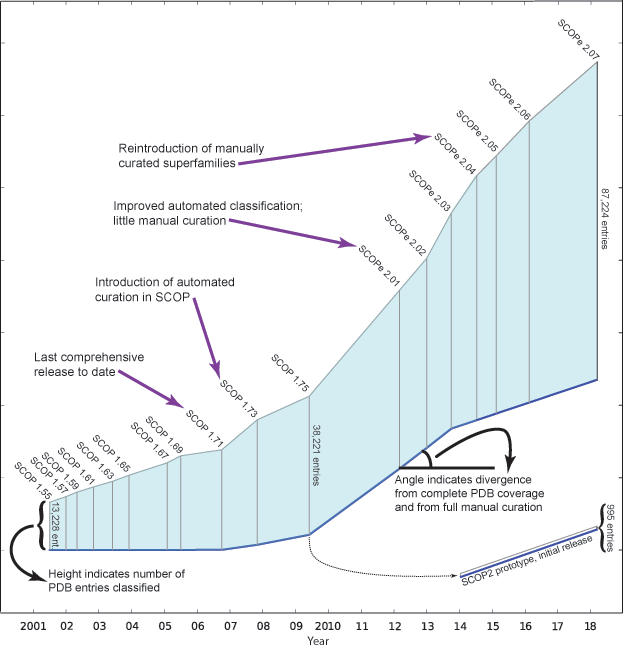
Note that since releases beyond SCOP 1.71 are not comprehensive, not all structurally characterized protein families and folds from the PDB are classified in these releases. Therefore, we caution against using later releases to (for example) analyze the rate at which new folds are being discovered.
What is the relationship between SCOP, SCOPe, and SCOP2?
- Applies to: SCOPe version 2.01 through current release
- References: 1-5
SCOP (version 1) is a database that originated in 1994 for protein structure classification [PubMed]. The last version of this database, 1.75, was released in June 2009 and is still available both in Cambridge UK and on the SCOPe website. SCOP 1.75 classifies 38,221 PDB entries.
SCOPe continues the development and maintenance of the classic SCOP hierarchy and may be seen as a continuation of the SCOP database, with ongoing updates and new classifications [PubMed]. SCOPe is completely backward compatible with SCOP (version 1), including all the same parseable files that we previously released separately in SCOP and Astral releases. As of March 2018, SCOPe 2.07-stable classifies 87,224 PDB entries.
SCOP2 represents a marked change from the SCOP classification structure and is currently available as a prototype with only a small number of classified families. SCOP2 replaces the tree-like SCOP hierarchy with a directed acyclic graph, allowing curators to represent structural and evolutionary relationships between proteins in more precise detail. There is no division of proteins into domains with fixed boundaries; instead, node-specific bounaries define the protein regions for each particular relationship [PubMed]. As of March 2018, the SCOP2 prototype classifies 995 PDB entries.
Stable identifiers
- Applies to: SCOP version 1.55 through current release
- Reference: 8
The SCOPe database continues to support the same style of stable identifiers in use since SCOP 1.55. Identifiers are provided as an unambiguous way to link to each a SCOP or SCOPe entry and are stable across releases. See the sections on citing and linking to SCOPe and SCOP using stable identifiers.
-
sccs. SCOP(e) concise classification string. This is a dot notation used to concisely describe a SCOP(e) class, fold, superfamily, and family. For example, a.39.1.1 references the "Calbindin D9K" family, where "a" represents the class, "39" represents the fold, "1" represents the superfamily, and the last "1" represents the family.
-
sunid. SCOP(e) unique identifier. This is simply a number that may be used to reference any entry in the SCOP(e) hierarchy, from root to leaves (Fold, Superfamily, Family, etc.).
-
sid. Stable domain identifier. A 7-character sid consists of "d" followed by the 4-character PDB ID of the file of origin, the PDB chain ID ('_' if none, '.' if multiple as is the case in genetic domains), and a single character (usually an integer) if needed to specify the domain uniquely ('_' if not). Sids are currently all lower case, even when the chain letter is upper case. Example sids include d4akea1, d9hvpa_, and d1cph.1.
Both sunids and sccs identifiers are expected to remain stable across releases, except in cases where the classification changes substantially. For example, when nodes in the hierarchy (e.g. Superfamilies) are merged or split due to new evidence of evolutionary relationships, corresponding identifiers become obsolete and new sunids are introduced. If a domain is split, or the boundaries change substantially, new sid(s) and sunid(s) are assigned.
A history of changes between all consecutive SCOP(e) releases is available under the Stats & History tab, and a history of changes to each individual entry is shown at the bottom of the page describing that entry.
Astral documentation
- Applies to: SCOP version 1.55 through current release
- References: 11-13
The primary sources of Astral documention are the three references cited above and listed on the Help > References page. The Astral compendium is a collection of software and databases, partially derived from SCOP(e), that aid research into protein structure and evolution. Astral provides sequences and coordinate files for all SCOP(e) domains, as well as sequences for all PDB chains that are classified in SCOP(e). Chemically modified amino acids are translated back to the original sequence, and sequences are curated to eliminate errors resulting from the automated parsing of PDB files. Because the majority of sequences in the PDB are very similar to others, Astral provides representative subsets of proteins that span the set of classified protein structures or domains while alleviating bias toward well-studied proteins. The highest quality representative in each subset is chosen using AEROSPACI scores, which provide a numeric estimate of the quality and precision of crystallographically determined structures.
The figure below, taken from the
2004 NAR paper, gives a brief overview
of how Astral is created. New releases
of Astral are derived from SCOPe domain definitions.

Data flow in Astral: Primary data sources are shown in green. Primary Astral databases are shown in light yellow. Less commonly used resources are shown in darker yellow. Resources added more recently are outlined in light blue/grey. Using the RAF maps, four complete sequence sets are created for every domain in the first seven classes of the SCOP(e) database. Two sets (the genetic domain sets) include the genetic domain sequences described in the 2002 NAR paper, and the other two (the original-style sequence sets) use the prior method of splitting each multi-chain domain into multiple sequences. For each of these methodologies, one complete sequence set is derived from sequences in the PDB ATOM records, and another from sequences in the SEQRES records. The SEQRES sets (for both genetic domain and original-style methods) are used to derive representative subsets. Each set is fully compared against itself using BLAST, and subsets are created using three similarity criteria and various thresholds. Representatives are chosen according to AEROSPACI scores. PDB chain sequence sets are derived from the SEQRES records of every PDB chain in SCOP(e); selected subsets are created at 90-100% ID thresholds. PDB-style files are derived from the RAF maps and SCOP(e) domain definitions. At each new release of Astral, all non-redundant sequences from each SCOP(e) family and superfamily are aligned using MAFFT. A hidden Markov model (HMM) is created from the multiple sequence alignment for each family and superfamily using HMMER. These HMMs, and BLAST, are used to predict domains in the sequences of newly released PDB entries on a weekly basis. HMMs from the Pfam-A database are also used to predict domains in regions of the sequences not identified by HMMs derived from Astral. Unassigned regions of at least 20 consecutive residues are also predicted to be potential domains.
Documentation describing more recent updates (since the 2004 paper) can be found in the release notes, which are linked from the Downloads > Astral Sequences & Subsets page.
Searching SCOP and SCOPe
- Applies to: SCOP version 1.55 through current release
- Reference: 8
The website search bar at the top of each page supports keyword search, as well as searches by a number of different SCOP(e) identifiers.
View the Search examples page for further details on the syntax, as well as many examples.
Domain visualization
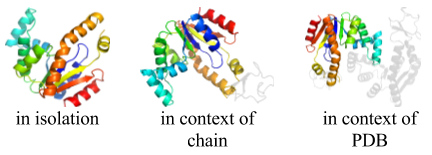
- Applies to: SCOP version 1.55 through current release
- Reference: 1
Thumbnails were generated for each domain using PyMOL, based on a viewing angle for each protein calculated using OVOP (Sverud O, MacCallum RM. 2003. Towards optimal views of proteins. Bioinformatics 19(7): 882-888 [PubMed]).
On the SCOPe website page showing information about each domain, thumbnails are displayed showing the domain in isolation, in the context of its chain, and in the context of its PDB structure. Links to other domains in the same chain, and in the same PDB structure, are below the corresponding thumbnail. To preview thumbnails from these other domains and get tool tip text with a short description, mouse over the links to the other domains. The figure to the right shows the thumbnails generated for the domain d4akea1.
Domain pages also include a JavaScript-based viewer that allows users to view and rotate domains in 3D without installing additional software. The 3D visualization domain visualization tool was built using JSmol, a Javascript-based viewer created by the Jmol project.
How to link to SCOP and SCOPe nodes
- Applies to: SCOP version 1.55 through current release
- References: 1,8
Because the SCOPe website is based on different technology from the older SCOP websites, old search paths (e.g., those using the search.cgi engine) are deprecated. URLs in the older style will be redirected approprately, so all old links to the Berkeley SCOP mirror should still work, including links to retrieve data that used to be on the Astral website (e.g., getseqs.cgi, spaci.cgi). We also support old-style links to all versions of SCOP with stable identifiers; for example, http://scop.berkeley.edu/scop/ links to SCOP 1.75, the last release of SCOP (version 1), while http://scop.berkeley.edu/scop-1.67/ links to SCOP version 1.67.
When making new links to SCOP or SCOPe data on this website, please use the new link formats below:
-
To link to a node the current release, use any
stable identifier (sunid,
sid or sccs).
The link formats are:
http://scop.berkeley.edu/sunid=xxxxx http://scop.berkeley.edu/sid=xxxxx http://scop.berkeley.edu/sccs=xxxxx
Linking by sunid is preferred because it is faster (sunid is a number, e.g. 46456; sids and sccs are both alphanumeric). Note that if the identifier you are linking to has become obsolete in the current release, the last release in which the identifier appears is shown, along with an explanation. -
To link to a node in a particular SCOP
or SCOPe
release, use the version number and sunid. The link format is:
http://scop.berkeley.edu/sunid=xxxxx&ver=yyyy
(versions are alphanumeric; e.g., 1.55 or 2.01).
You can also use this style of linking to link to nodes by sid or by sccs. -
To link to the root of a particular SCOP
or SCOPe
release, use the version number. The link format is:
http://scop.berkeley.edu/ver=yyyy
(versions are alphanumeric; e.g., 1.55 or 2.01). -
To link to information we have on a PDB
file, use its PDB code. The link
format is:
http://scop.berkeley.edu/pdb/code=xxxx
We plan for this link format to remain stable for all future versions of SCOPe unless there is a major change to the classification, even if we make changes to our underlying website technology.
How to cite SCOP and SCOPe entries
- Applies to: SCOP version 1.55 through current release
- References: 1,8
In the interest of scientific reproducibility, when you publish a manuscript or give a talk about data you obtained from SCOP or SCOPe, it's crucial to unambiguously refer to the data you used. Using these citation conventions will provide all the information needed for other researchers to accurately download the same data that you are working with. For related help, see sections on backward compatibility with SCOP, stable releases and updates, and stable identifiers.
When citing a particular SCOPe or SCOP entry, we recommend the following conventions, depending on the level of the entry cited:
- Class: Use the complete SCOP(e)
version number, full name of the class, and
sunid. Examples:
- SCOPe 2.03-stable Class a: All alpha proteins [46456]
- SCOP 1.67 Class c: Alpha and beta proteins (a/b) [51349]
- Fold, Superfamily, or Family: Use the complete SCOP(e)
version number, sccs,
full name, and sunid. Examples:
- SCOPe 2.03-2013-12-12 Fold b.1: Immunoglobulin-like beta-sandwich [48725]
- SCOPe 2.03-stable Family c.2.1.1: Alcohol dehydrogenase-like, C-terminal domain [51736]
-
Protein or Species: Use the complete SCOP(e)
version number, full
name, sunid, and sccs. Examples:
- SCOPe 2.03-stable Protein Adenylate kinase [52554] (sccs c.37.1.1)
- SCOP 1.75 Species: Baker's yeast (Saccharomyces cerevisiae) [TaxId: 4932] [56639] (sccs e.5.1.1)
-
Domain. Use the complete SCOP(e)
version number, sid, sunid, and sccs. Examples:
- SCOPe 2.03-stable d1p0ca2 [87639] (sccs c.2.1.1)
- SCOPe 2.03-2013-12-12 d4cadj2 [229759] (sccs b.1.1.0)
Publications using SCOP, Astral or SCOPe data should also cite the appropriate references given on the Help > References page rather than just referring to the website.
Parseable files
- Applies to: SCOP version 1.55 through current release
- Reference: 8
Parseable files have been provided for each SCOP(e) release, available from the Downloads > Parseable Files & Software page. Each of these files has a header, starting with the '#' character, which includes release, version, and copyright information.
The list below shows the four parseable formats for SCOP(e) data. For information on downloading and parsing Astral subsets and RAF sequence maps, see the Astral help.
-
dir.des.scop(e).txt. - Descriptions for each node in the SCOP(e) hierarchy.
46457 cf a.1 - Globin-like 113449 px a.1.1.1 d1ux8a_ 1ux8 A: ^1 ^2 ^3 ^4 ^5
Five tab-delimited columns:
- sunid
- level: cl - class; cf - fold; sf - superfamily; fa - family; dm - protein; sp - species; px - domain
- sccs
- sid or "-"
- description
-
dir.cla.scop(e).txt - Full classification for each domain.
d1ux8a_ 1ux8 A: a.1.1.1 113449 cl=46456,cf=46457,sf=46458,fa=46459,dm=46460,sp=116748,px=113449 2gtld1 2gtl D:8-147 a.1.1.2 135658 cl=46456,cf=46457,sf=46458,fa=46463,dm=116758,sp=116759,px=135658 d1cph.1 1cph B:,A: g.1.1.1 43831 cl=56992,cf=56993,sf=56994,fa=56995,dm=56996,sp=56997,px=43831 ^1 ^2 ^3 ^4 ^5 ^6
Six tab-delimited columns:
- sid
- PDB ID
- description
- sccs
- sunid
- sunids of ancestor nodes in comma-delimited list, in the format "level=sunid". For level codes, see description for dir.des.scop(e).txt.
-
dir.hie.scop(e).txt - Children and parents for each node
0 - 46456,48724,51349,53931,56572,56835,56992,57942,58117,58231,58788 46457 46456 46458,46548 ^1 ^2 ^3
Three tab-delimited columns:
- sunid
- parent's sunid or "-"
- comma-separated list of children's sunids or "-"
-
dir.com.scop(e).txt - Human and machine-annotated comments
100067 ! complexed with cyn, hem, xe 63437 ! SQ Q10784 164742 ! automated match to d1s56b_ ! complexed with cyn, hem, na; mutant ^1 ^2 ^3
Two or more "!"-delimited columns:
- sunid
- first comment
- additional comments (one per column)
Backward compatibility with SCOP
- Applies to: SCOPe version 2.01 through current release
- Reference: 1
In order to facilitate use of SCOPe data by SCOP and Astral users, we provide SCOPe data in parseable files in the same formats as the SCOP and Astral databases. SCOPe uses the same stable identifiers (e.g., sunid, sid, sccs) as were used for prior releases of SCOP and Astral, and the same protocols previously used to assign new SCOP and Astral identifiers are also used in SCOPe. A history of all changes between consecutive releases of SCOP and SCOPe is available on the Stats & History page.
Automatic classification of PDB entries
- Applies to: SCOP version 1.55 through SCOP version 1.73
- Reference: 14
The vast majority of new protein structures solved represent a new experiment on a protein that was already structurally characterized (Chandonia and Brenner, 2006, available on the Help > References page). Our goal in implementing automation is classify structures that can be reliably classified in the existing SCOP hierarchy, without sacrificing the reliability of the database, which has been maintained through years of expert curation.
The Astral authors have been provisionally classifying new PDB structures using the ASTEROIDS procedure, explained below and documented in the Chandonia et. al., 2004 paper (available on the Help > References page). We benchmarked the ASTEROIDS algorithm against all stable versions of SCOP from 1.55 to 1.75 using the method described in the Benchmarking in order to determine whether a subset of the predictions were 100% reliable.
To fully classify domain predictions in the SCOPe hierarchy, we also had to assign levels below Superfamily. In our benchmarking, we found that the following rules could be used to reliably assign those levels:
- Family can be assigned if the BLAST percent identity to the previously classified domain is at least 55%, or if the gene annotation provided in the PDB file matched the gene annotation of exactly one family in the assigned Superfamily.
- Protein can be assigned if the BLAST percent identity to the previously classified domain is at least 99%, or if the gene annotation provided in the PDB file matched the gene annotation of exactly one protein in the assigned Family. This relatively high threshold is required because some paralogs in SCOP have ~98% identity to each other, and are classified as separate proteins.
- Species can not be reliably assigned based on sequence similarity, but can often be matched to a species in SCOP based on the species annotation in the PDB file.
In cases where the protein or family could not be reliably matched to an existing SCOP clade, we created a new group called "automated matches" at the Family or Protein level. These are not true families or proteins, because the domains within each one are not as similar to each other as members of manually curated families and proteins. For example, assignment to an "automated matches" family means the domain could reliably be assigned to the parent superfamily, but could not be automatically assigned to one of the existing families with 100% confidence. Manual curation or future improvements in automation may allow us to re-assign these domains with greater specificity. New SCOPe Species nodes were created if the PDB species annotation did not match any existing SCOP species.
To facilitate the mapping of SCOPe Species nodes to PDB species annotations, we attempted to make the descriptions of SCOPe species-level nodes more consistent, e.g., by always using the same common name for a species, and by updating NCBI taxonomy ids that have changed. This resulted in 1,520 species nodes being updated between SCOP 1.75 and SCOPe 2.01. The sunids for these nodes did NOT change, since each refers to the same species as before.
We found that ASTEROIDS predictions that met all of the following criteria were as reliable as manually curated domains (i.e., no errors that were revealed by manual inspection):
- The ASTEROIDS prediction was based on BLAST, with an E-value at least as significant as 0.0001.
- The ASTEROIDS prediction spanned the entire PDB chain.
- The BLAST hit spanned the entire domain, with no gaps longer than 10 residues.
- The domain hit by BLAST spanned its entire PDB chain.
- The predicted domain was in the first 7 SCOP classes.
- In cases where the predicted ASTEROIDS boundaries were within 20 residues of the end of the chain, we extended them to include the entire chain. This prevents domains from being truncated in cases where a newly solved domain is slightly longer than a previously characterized domain.
- The predicted domain had a resolution below 3.0 Å, and was not annotated by the PDB as synthetic or a ribosome. This prevents domains from being incorrectly matched to the first 7 SCOP classes, in cases where manual curation had put the domain in classes 8 and above.
Examination of all differences in these predictions revealed 2 errors in domains that had been manually curated for prior versions of SCOP, as well as several truncation errors in domains that had been automatically classified for prior versions of SCOP. We also noticed a number of previously automatically classified domains that were classified incorrectly at the Species or Protein levels. Errors were corrected in SCOPe 2.01, and changes are listed in the Stats & History page.
Using the new automation procedure, we:
- Added 11,152 PDB files to SCOPe 2.01 that were not classified in SCOP 1.75.
- Replaced old automatically assigned domains, in cases where an updated prediction was available. This resulted in the reannotation of 10,421 domains.
- Corrected 58 erroneous protein/species annotations (see history) and re-assigned all the replaced domains above (and all the new domains) to the most specific level we could confidently assign.
Benchmarking
- Applies to: SCOPe version 2.01 through current release
- Reference: 1
To validate the automatic classification method, we performed benchmarking against all SCOP releases with stable identifiers (i.e. releases 1.55-1.75). All PDB entries that were added between each pair of consecutive releases were automatically classified based on the earlier release and compared with the manually curated domains in the subsequent release. A predicted domain was considered identified and classified correctly if:
- Was classified in the same Superfamily as the manually curated domain.
- Had the same number of regions (i.e., consecutive parts of the sequence) as the manually curated domain.
- Had all region boundaries within 10 residues of the manually curated boundaries.
We have taken a very conservative approach to applying automated classification, in order to ensure that our methods are as accurate as manual curation, even though there are many proteins to which our methods cannot be applied. This means that our automated methods were tuned until they produced zero errors relative to manual curation.
Plans for future website development
We are continuing to upgrade the website. Planned features include more sophisticated search functionality (including searches by sequence and by structure), use of AJAX to avoid full page reloads, and a SCOPe tree browser for more efficient display of results. We also plan to allow downloads of additional derived data (e.g., sequence sets and HMMs) for different clades in the tree.
Further help
The primary sources of documentation are the papers listed on the Help > References page.Please report bugs and usability issues to us at scope@compbio.berkeley.edu.
