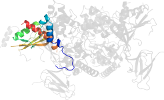
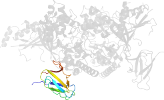
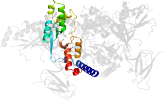
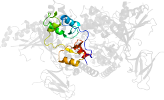
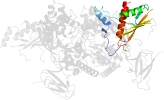
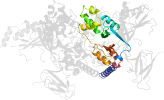
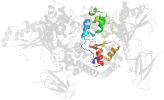
Lineage for d2qlve2 (2qlv E:307-412)
- Root: SCOPe 2.06

Class d: Alpha and beta proteins (a+b) [53931] (385 folds) 
Fold d.353: AMPKBI-like [160218] (1 superfamily)
comprises 3 short helices and 3-stranded meander beta-sheet; makes extensive intermolecular interactions
Superfamily d.353.1: AMPKBI-like [160219] (1 family) 
automatically mapped to Pfam PF04739
Family d.353.1.1: AMPKBI-like [160220] (4 proteins)
Pfam PF04739; 5'-AMP-activated protein kinase, beta subunit, complex-interacting region
Protein SIP2 [160221] (1 species) 
Species Baker's yeast (Saccharomyces cerevisiae) [TaxId:4932] [160222] (1 PDB entry)
Uniprot P34164 306-412
Domain d2qlve2: 2qlv E:307-412 [150871]
Other proteins in same PDB: d2qlva1, d2qlvb1, d2qlvc1, d2qlvc2, d2qlvd_, d2qlve1, d2qlvf1, d2qlvf2
automated match to d2qlvb2
Details for d2qlve2
PDB Entry: 2qlv (more details), 2.6 Å
PDB Description: crystal structure of the heterotrimer core of the s. cerevisiae ampk homolog snf1
PDB Compounds: (E:) Protein SIP2SCOPe Domain Sequences for d2qlve2:
Sequence, based on SEQRES records: (download)
>d2qlve2 d.353.1.1 (E:307-412) SIP2 {Baker's yeast (Saccharomyces cerevisiae) [TaxId: 4932]}
tdipavftdpsvmeryyytldrqqsntdtswltppqlppqlenvilnkyyatqdqfnenn
sgalpipnhvvlnhlvtssikhntlcvasivrykqkyvtqilytpi
Sequence, based on observed residues (ATOM records): (download)
>d2qlve2 d.353.1.1 (E:307-412) SIP2 {Baker's yeast (Saccharomyces cerevisiae) [TaxId: 4932]}
tdipavftppqlppqhvvlnhlvtssikhntlcvasivrykqkyvtqilytpi
SCOPe Domain Coordinates for d2qlve2:
Click to download the PDB-style file with coordinates for d2qlve2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d2qlve2:
- d2qlve2 first appeared in SCOP 1.75
- d2qlve2 appears in SCOPe 2.05
- d2qlve2 appears in SCOPe 2.07
- d2qlve2 appears in the current release, SCOPe 2.08

 View in 3D
View in 3D