Lineage for d1xac__ (1xac -)
- Root: SCOP 1.69

Class c: Alpha and beta proteins (a/b) [51349] (136 folds) 
Fold c.77: Isocitrate/Isopropylmalate dehydrogenase-like [53658] (1 superfamily)
consists of two intertwined (sub)domains related by pseudo dyad; duplication
3 layers: a/b/a; single mixed beta-sheet of 10 strands, order 213A945867 (A=10); strands from 5 to 9 are antiparallel to the rest
Superfamily c.77.1: Isocitrate/Isopropylmalate dehydrogenase-like [53659] (5 families) 
the constituent families form similar dimers
Family c.77.1.1: Dimeric isocitrate & isopropylmalate dehydrogenases [53660] (3 proteins)
the active site is between the two identical subunits
Protein 3-isopropylmalate dehydrogenase, IPMDH [53661] (7 species) 
Species Chimera (Thermus thermophilus) and (Bacillus subtilis) [TaxId:274] [53663] (2 PDB entries) 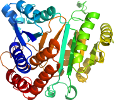
Domain d1xac__: 1xac - [35056]
Details for d1xac__
PDB Entry: 1xac (more details), 2.1 Å
PDB Description: chimera isopropylmalate dehydrogenase between bacillus subtilis (m) and thermus thermophilus (t) from n-terminal: 20% t middle 20% m residual 60% t, mutated at s82r. low temperature (100k) structure.
SCOP Domain Sequences for d1xac__:
Sequence; same for both SEQRES and ATOM records: (download)
>d1xac__ c.77.1.1 (-) 3-isopropylmalate dehydrogenase, IPMDH {Chimera (Thermus thermophilus) and (Bacillus subtilis)}
mkvavlpgdgigpevteaalkvlraldeaeglglayevfpfggaaidafgepfpeptrkg
veeaeavllgsvggpkwdqnprelrpekgllsirkqldlfanlrpvkvfeslsdasplkk
eyidnvdfvivreltggiyfgeprgmseaeawnteryskpevervarvafeaarkrrkhv
vsvdkanvlevgefwrktveevgrgypdvalehqyvdamamhlvrsparfdvvvtgnifg
dilsdlasvlpgslgllpsaslgrgtpvfepvhgsapdiagkgianptaailsaammleh
afglvelarkvedavakalletpppdlggsagteaftatvlrhla
SCOP Domain Coordinates for d1xac__:
Click to download the PDB-style file with coordinates for d1xac__.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1xac__:
- d1xac__ first appeared (with stable ids) in SCOP 1.55
- d1xac__ appears in SCOP 1.67
- d1xac__ appears in SCOP 1.71
- d1xac__ appears in the current release, SCOPe 2.08, called d1xaca_
 View in 3D
View in 3D