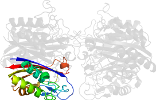
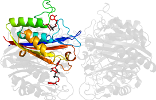
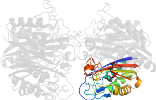
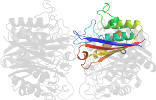
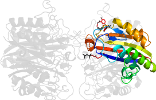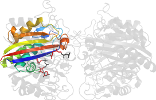Lineage for d2amtd_ (2amt D:)
- Root: SCOPe 2.05

Class d: Alpha and beta proteins (a+b) [53931] (381 folds) 
Fold d.79: Bacillus chorismate mutase-like [55297] (9 superfamilies)
core: beta-alpha-beta-alpha-beta(2); mixed beta-sheet: order: 1423, strand 4 is antiparallel to the rest
Superfamily d.79.5: IpsF-like [69765] (2 families) 
forms trimers with three closely packed beta-sheets; possible link between the YjgF-like (d.79.1) and 4'-phosphopantetheinyl transferase superfamilies (d.150.1)
Family d.79.5.1: IpsF-like [69766] (3 proteins)
automatically mapped to Pfam PF02542
Protein 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase IspF [69767] (6 species) 
Species Escherichia coli [TaxId:562] [69768] (13 PDB entries)
Uniprot P62617 ! Uniprot P36663
Domain d2amtd_: 2amt D: [162837]
automated match to d1h48c_
complexed with 1aa, gpp, zn
Details for d2amtd_
PDB Entry: 2amt (more details), 2.3 Å
PDB Description: Structure of 2C-Methyl-D-Erythritol 2,4-Clycodiphosphate Synthase complexed with a CDP derived fluorescent inhibitor
PDB Compounds: (D:) 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthaseSCOPe Domain Sequences for d2amtd_:
Sequence; same for both SEQRES and ATOM records: (download)
>d2amtd_ d.79.5.1 (D:) 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase IspF {Escherichia coli [TaxId: 562]}
mrighgfdvhafggegpiiiggvripyekgllahsdgdvalhaltdallgaaalgdigkl
fpdtdpafkgadsrellreawrriqakgytlgnvdvtiiaqapkmlphipqmrvfiaedl
gchmddvnvkattteklgftgrgegiaceavallik
SCOPe Domain Coordinates for d2amtd_:
Click to download the PDB-style file with coordinates for d2amtd_.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d2amtd_:
- d2amtd_ first appeared in SCOPe 2.01
- d2amtd_ appears in SCOPe 2.04
- d2amtd_ appears in SCOPe 2.06
- d2amtd_ appears in the current release, SCOPe 2.08
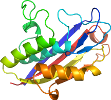
 View in 3D
View in 3D