Lineage for d1kfva3 (1kfv A:227-271)
- Root: SCOPe 2.08
Class g: Small proteins [56992] (100 folds) 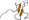
Fold g.39: Glucocorticoid receptor-like (DNA-binding domain) [57715] (1 superfamily)
alpha+beta metal(zinc)-bound fold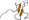
Superfamily g.39.1: Glucocorticoid receptor-like (DNA-binding domain) [57716] (19 families) 
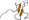
Family g.39.1.8: C-terminal, Zn-finger domain of MutM-like DNA repair proteins [81627] (3 proteins) 
Protein DNA repair protein MutM (Fpg) [81622] (4 species) 
Species Lactococcus lactis [TaxId:1358] [81619] (7 PDB entries)
Uniprot P42371
Domain d1kfva3: 1kfv A:227-271 [75888]
Other proteins in same PDB: d1kfva1, d1kfva2, d1kfvb1, d1kfvb2
protein/DNA complex; complexed with gol, zn
Details for d1kfva3
PDB Entry: 1kfv (more details), 2.55 Å
PDB Description: crystal structure of lactococcus lactis formamido-pyrimidine dna glycosylase (alias fpg or mutm) non covalently bound to an ap site containing dna.
PDB Compounds: (A:) Formamido-pyrimidine DNA glycosylaseSCOPe Domain Sequences for d1kfva3:
Sequence; same for both SEQRES and ATOM records: (download)
>d1kfva3 g.39.1.8 (A:227-271) DNA repair protein MutM (Fpg) {Lactococcus lactis [TaxId: 1358]}
stgkmqnelqvygktgekcsrcgaeiqkikvagrgthfcpvcqqk
SCOPe Domain Coordinates for d1kfva3:
Click to download the PDB-style file with coordinates for d1kfva3.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1kfva3:
- d1kfva3 first appeared in SCOP 1.63
- d1kfva3 appears in SCOPe 2.07
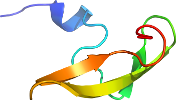
 View in 3D
View in 3D