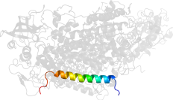
Lineage for d1eysh2 (1eys H:7-43)
- Root: SCOP 1.57

Class f: Membrane and cell surface proteins and peptides [56835] (11 folds) 
Fold f.2: Membrane all-alpha [56868] (1 superfamily) 
Superfamily f.2.1: Membrane all-alpha [56869] (10 families) 

Family f.2.1.2: Photosynthetic reaction centre, L-, M- and H-chains [56878] (1 protein) 
Protein Photosynthetic reaction centre, L-, M- and H-chains [56879] (3 species) 
Species Thermochromatium tepidum [TaxId:1050] [56882] (1 PDB entry) 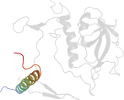
Domain d1eysh2: 1eys H:7-43 [43517]
Other proteins in same PDB: d1eysc_, d1eysh1
Details for d1eysh2
PDB Entry: 1eys (more details), 2.2 Å
PDB Description: crystal structure of photosynthetic reaction center from a thermophilic bacterium, thermochromatium tepidum
SCOP Domain Sequences for d1eysh2:
Sequence; same for both SEQRES and ATOM records: (download)
>d1eysh2 f.2.1.2 (H:7-43) Photosynthetic reaction centre, L-, M- and H-chains {Thermochromatium tepidum}
hyidaaqitiwafwlfffgliiylrredkregyplds
SCOP Domain Coordinates for d1eysh2:
Click to download the PDB-style file with coordinates for d1eysh2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1eysh2:

 View in 3D
View in 3D