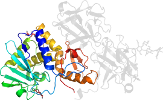
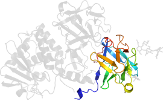Lineage for d4zfwa_ (4zfw A:)
- Root: SCOPe 2.06

Class d: Alpha and beta proteins (a+b) [53931] (385 folds) 
Fold d.165: Ribosome inactivating proteins (RIP) [56370] (1 superfamily)
contains mixed beta-sheet
Superfamily d.165.1: Ribosome inactivating proteins (RIP) [56371] (3 families) 

Family d.165.1.0: automated matches [191615] (1 protein)
not a true family
Protein automated matches [191124] (7 species)
not a true protein
Species Momordica charantia [TaxId:3673] [312311] (9 PDB entries) 
Domain d4zfwa_: 4zfw A: [313038]
Other proteins in same PDB: d4zfwb1, d4zfwb2
automated match to d1abra_
complexed with bma, fuc, gal, gla, gol, nag, xyp
Details for d4zfwa_
PDB Entry: 4zfw (more details), 2.54 Å
PDB Description: structural studies on a non-toxic homologue of type ii rips from momordica charantia (bitter gourd) in complex with galactose.
PDB Compounds: (A:) rRNA N-glycosidaseSCOPe Domain Sequences for d4zfwa_:
Sequence; same for both SEQRES and ATOM records: (download)
>d4zfwa_ d.165.1.0 (A:) automated matches {Momordica charantia [TaxId: 3673]}
nlslsqsnfsadtyksfiknlrkqltigasygsagipilkhsvpicerfllvdltngdne
titlainvedagfaayraadrsyffqnappiasyviftdtnqnimnfnntfesieivggt
trsetplgimhfeasifhlfvhdenyvptsflvliqmvleaakfkfieqkvihsimdmed
ftpglamlsleenwtqlslqlqaseslngvfgdsvslynsmdepigvdsmyypiltanma
fqlyqcp
SCOPe Domain Coordinates for d4zfwa_:
Click to download the PDB-style file with coordinates for d4zfwa_.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d4zfwa_:
- d4zfwa_ is new in SCOPe 2.06-stable
- d4zfwa_ appears in SCOPe 2.07
- d4zfwa_ appears in the current release, SCOPe 2.08
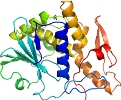
 View in 3D
View in 3D