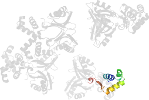Lineage for d2znzb2 (2znz B:85-170)
- Root: SCOPe 2.08

Class d: Alpha and beta proteins (a+b) [53931] (396 folds) 
Fold d.58: Ferredoxin-like [54861] (62 superfamilies)
alpha+beta sandwich with antiparallel beta-sheet; (beta-alpha-beta)x2
Superfamily d.58.4: Dimeric alpha+beta barrel [54909] (24 families) 
dimerizes through the beta-sheet; forms beta-sheet barrel, closed (n=8, S=12); dimers may assemble in higher oligomers
Family d.58.4.2: Lrp/AsnC-like transcriptional regulator C-terminal domain [69733] (5 proteins)
octamer: tetramer of dimers
automatically mapped to Pfam PF01037
Protein Putative transcriptional regulator PH1519 [102968] (1 species)
archaeal feast/famine regulatory protein
Species Pyrococcus horikoshii [TaxId:53953] [102969] (4 PDB entries)
identical sequence to Pyrococcus sp. ot3 protein
Domain d2znzb2: 2znz B:85-170 [207919]
Other proteins in same PDB: d2znza1, d2znzb1, d2znzc1, d2znzd1, d2znze1, d2znzf1, d2znzg1, d2znzh1
automated match to d1ri7a2
complexed with lys
Details for d2znzb2
PDB Entry: 2znz (more details), 2.39 Å
PDB Description: Crystal structure of FFRP
PDB Compounds: (B:) Uncharacterized HTH-type transcriptional regulator PH1519SCOPe Domain Sequences for d2znzb2:
Sequence; same for both SEQRES and ATOM records: (download)
>d2znzb2 d.58.4.2 (B:85-170) Putative transcriptional regulator PH1519 {Pyrococcus horikoshii [TaxId: 53953]}
ysmlafilvkvkagkysevasnlakypeivevyettgdydmvvkirtknseelnnfldli
gsipgvegthtmivlkthkettelpi
SCOPe Domain Coordinates for d2znzb2:
Click to download the PDB-style file with coordinates for d2znzb2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d2znzb2:
- d2znzb2 first appeared in SCOPe 2.03
- d2znzb2 appears in SCOPe 2.07
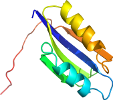
 View in 3D
View in 3D