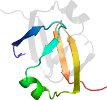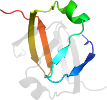Lineage for d1yfba1 (1yfb A:3-53)
- Root: SCOP 1.73

Class b: All beta proteins [48724] (165 folds) 
Fold b.129: Double-split beta-barrel [89446] (2 superfamilies)
pseudobarrel; capped on both ends by alpha-helices
Superfamily b.129.1: AbrB/MazE/MraZ-like [89447] (3 families) 
members of this superfamily are known or predicted to have DNA-binding function
Family b.129.1.3: Transcription-state regulator AbrB, the N-terminal DNA recognition domain [54743] (1 protein)
dimeric fold similar to MazE; the DNA-binding site is similar to the conserved surface site of MraZ
Protein Transcription-state regulator AbrB, the N-terminal DNA recognition domain [54744] (1 species) 
Species Bacillus subtilis [TaxId:1423] [54745] (3 PDB entries) 
Domain d1yfba1: 1yfb A:3-53 [123057]
automatically matched to d1ekta_
Details for d1yfba1
PDB Entry: 1yfb (more details)
PDB Description: the solution structure of the n-domain of the transcription factor abrb
PDB Compounds: (A:) Transition state regulatory protein abrBSCOP Domain Sequences for d1yfba1:
Sequence; same for both SEQRES and ATOM records: (download)
>d1yfba1 b.129.1.3 (A:3-53) Transcription-state regulator AbrB, the N-terminal DNA recognition domain {Bacillus subtilis [TaxId: 1423]}
mkstgivrkvdelgrvvipielrrtlgiaekdaleiyvddekiilkkykpn
SCOP Domain Coordinates for d1yfba1:
Click to download the PDB-style file with coordinates for d1yfba1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1yfba1:
- d1yfba1 is new in SCOP 1.73
- d1yfba1 appears in SCOP 1.75
- d1yfba1 appears in the current release, SCOPe 2.08, called d1yfba_

 View in 3D
View in 3D