Lineage for d1yc2e_ (1yc2 E:)
- Root: SCOPe 2.08

Class c: Alpha and beta proteins (a/b) [51349] (148 folds) 
Fold c.31: DHS-like NAD/FAD-binding domain [52466] (1 superfamily)
3 layers: a/b/a; parallel beta-sheet of 6 strands, order 321456; Rossmann-like
Superfamily c.31.1: DHS-like NAD/FAD-binding domain [52467] (7 families) 
binds cofactor molecules in the opposite direction than classical Rossmann fold
Family c.31.1.5: Sir2 family of transcriptional regulators [63984] (8 proteins)
silent information regulator 2; contains insertion of a rubredoxin-like zinc finger domain
Protein AF0112, Sir2 homolog (Sir2-AF2) [82380] (1 species) 
Species Archaeoglobus fulgidus [TaxId:2234] [82381] (3 PDB entries) 
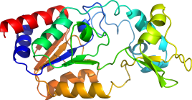
Domain d1yc2e_: 1yc2 E: [122907]
automated match to d1ma3a_
complexed with 2pe, apr, edo, nad, nca, pg4, pge, so4, zn
Details for d1yc2e_
PDB Entry: 1yc2 (more details), 2.4 Å
PDB Description: Sir2Af2-NAD-ADPribose-nicotinamide
PDB Compounds: (E:) NAD-dependent deacetylase 2SCOPe Domain Sequences for d1yc2e_:
Sequence; same for both SEQRES and ATOM records: (download)
>d1yc2e_ c.31.1.5 (E:) AF0112, Sir2 homolog (Sir2-AF2) {Archaeoglobus fulgidus [TaxId: 2234]}
medeirkaaeilakskhavvftgagisaesgiptfrgedglwrkydpeevasisgfkrnp
rafwefsmemkdklfaepnpahyaiaelermgivkavitqnidmlhqragsrrvlelhgs
mdkldcldchetydwsefvedfnkgeiprcrkcgsyyvkprvvlfgeplpqrtlfeaiee
akhcdafmvvgsslvvypaaelpyiakkagakmiivnaeptmadpifdvkiigkagevlp
kiveevkrlr
SCOPe Domain Coordinates for d1yc2e_:
Click to download the PDB-style file with coordinates for d1yc2e_.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1yc2e_:
- d1yc2e_ first appeared in SCOP 1.73, called d1yc2e1
- d1yc2e_ appears in SCOPe 2.07
 View in 3D View in 3DDomains from other chains: (mouse over for more information) d1yc2a_, d1yc2b_, d1yc2c_, d1yc2d_ |