Lineage for d1svia_ (1svi A:)
- Root: SCOPe 2.08

Class c: Alpha and beta proteins (a/b) [51349] (148 folds) 
Fold c.37: P-loop containing nucleoside triphosphate hydrolases [52539] (1 superfamily)
3 layers: a/b/a, parallel or mixed beta-sheets of variable sizes
Superfamily c.37.1: P-loop containing nucleoside triphosphate hydrolases [52540] (27 families) 
division into families based on beta-sheet topologies
Family c.37.1.8: G proteins [52592] (81 proteins)
core: mixed beta-sheet of 6 strands, order 231456; strand 2 is antiparallel to the rest
Protein Probable GTPase EngB [89667] (2 species) 
Species Bacillus subtilis [TaxId:1423] [110534] (3 PDB entries)
Uniprot P38424
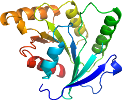
Domain d1svia_: 1svi A: [106048]
complexed with gdp
Details for d1svia_
PDB Entry: 1svi (more details), 1.95 Å
PDB Description: crystal structure of the gtp-binding protein ysxc complexed with gdp
PDB Compounds: (A:) GTP-binding protein YsxCSCOPe Domain Sequences for d1svia_:
Sequence, based on SEQRES records: (download)
>d1svia_ c.37.1.8 (A:) Probable GTPase EngB {Bacillus subtilis [TaxId: 1423]}
mkvtkseivisavkpeqypegglpeialagrsnvgkssfinslinrknlartsskpgktq
tlnfyiindelhfvdvpgygfakvsksereawgrmietyittreelkavvqivdlrhaps
nddvqmyeflkyygipviviatkadkipkgkwdkhakvvrqtlnidpedelilfssetkk
gkdeawgaikkminr
Sequence, based on observed residues (ATOM records): (download)
>d1svia_ c.37.1.8 (A:) Probable GTPase EngB {Bacillus subtilis [TaxId: 1423]}
mkvtkseivisavkpeqypegglpeialagrsnvgkssfinslinrqtlnfyiindelhf
vdvpgygfakvsksereawgrmietyittreelkavvqivdlrhapsnddvqmyeflkyy
gipviviatkadkipkgkwdkhakvvrqtlnidpedelilfssetkkgkdeawgaikkmi
nr
SCOPe Domain Coordinates for d1svia_:
Click to download the PDB-style file with coordinates for d1svia_.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1svia_:
- d1svia_ first appeared in SCOP 1.69
- d1svia_ appears in SCOPe 2.07
 View in 3D
View in 3D