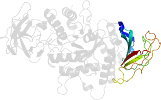
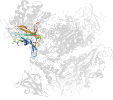
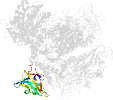
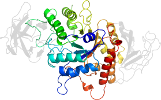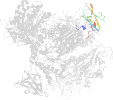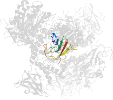Lineage for d1m7xa1 (1m7x A:117-226)
- Root: SCOPe 2.08

Class b: All beta proteins [48724] (180 folds) 
Fold b.1: Immunoglobulin-like beta-sandwich [48725] (33 superfamilies)
sandwich; 7 strands in 2 sheets; greek-key
some members of the fold have additional strands
Superfamily b.1.18: E set domains [81296] (27 families) 
"Early" Ig-like fold families possibly related to the immunoglobulin and/or fibronectin type III superfamilies
Family b.1.18.2: E-set domains of sugar-utilizing enzymes [81282] (21 proteins)
domains of unknown function associated with different type of catalytic domains in a different sequential location
subgroup of the larger IPT/TIG domain family
Protein 1,4-alpha-glucan branching enzyme, N-terminal domain N [81962] (1 species)
domain architecture similar to isoamylase
Species Escherichia coli [TaxId:562] [81963] (1 PDB entry) 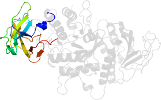
Domain d1m7xa1: 1m7x A:117-226 [78749]
Other proteins in same PDB: d1m7xa2, d1m7xa3, d1m7xb2, d1m7xb3, d1m7xc2, d1m7xc3, d1m7xd2, d1m7xd3
Details for d1m7xa1
PDB Entry: 1m7x (more details), 2.3 Å
PDB Description: the x-ray crystallographic structure of branching enzyme
PDB Compounds: (A:) 1,4-alpha-glucan Branching EnzymeSCOPe Domain Sequences for d1m7xa1:
Sequence; same for both SEQRES and ATOM records: (download)
>d1m7xa1 b.1.18.2 (A:117-226) 1,4-alpha-glucan branching enzyme, N-terminal domain N {Escherichia coli [TaxId: 562]}
thlrpyetlgahadtmdgvtgtrfsvwapnarrvsvvgqfnywdgrrhpmrlrkesgiwe
lfipgahngqlykyemidangnlrlksdpyafeaqmrpetaslicglpek
SCOPe Domain Coordinates for d1m7xa1:
Click to download the PDB-style file with coordinates for d1m7xa1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1m7xa1:
- d1m7xa1 first appeared in SCOP 1.63
- d1m7xa1 appears in SCOPe 2.07

 View in 3D
View in 3D