Lineage for Family b.1.18.2: E-set domains of sugar-utilizing enzymes
- Root: SCOPe 2.08

Class b: All beta proteins [48724] (180 folds) 
Fold b.1: Immunoglobulin-like beta-sandwich [48725] (33 superfamilies)
sandwich; 7 strands in 2 sheets; greek-key
some members of the fold have additional strands
Superfamily b.1.18: E set domains [81296] (27 families) 
"Early" Ig-like fold families possibly related to the immunoglobulin and/or fibronectin type III superfamilies
Family b.1.18.2: E-set domains of sugar-utilizing enzymes [81282] (21 proteins)
domains of unknown function associated with different type of catalytic domains in a different sequential location
subgroup of the larger IPT/TIG domain family
Proteins:

1,4-alpha-glucan branching enzyme, N-terminal domain N [81962] (1 species)
domain architecture similar to isoamylase
Species Escherichia coli [TaxId:562] [81963] (1 PDB entry)
Bacterial chitobiase (N-acetyl-beta-glucoseaminidase), C-terminal domain [49211] (1 species)
rudiment form of Ig-like domain; follows the catalytic (beta/alpha)8-barrel domain; family 20 glycosyl hydrolases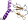
Species Serratia marcescens [TaxId:615] [49212] (4 PDB entries)

Beta-1,4-mannanase domain 2 (postcatalytic) [141020] (1 species) 
Species Cellulomonas fimi [TaxId:1708] [141021] (2 PDB entries)
Uniprot Q9XCV5 421-514

Beta-galactosidase [419186] (1 species)
Pfam PF16355
inserted beta hairpin between strands 4 and 5 of canonical fold
Species Streptococcus pneumoniae TIGR4 [TaxId:170187] [419697] (3 PDB entries)

CelD cellulase, N-terminal domain [49231] (1 species)
precedes the catalytic alpha6/alpha6 domain
Species Clostridium thermocellum [TaxId:1515] [49232] (1 PDB entry)

Cellulose 1,4-beta-cellobiosidase CbhA, precatalytic domain [101521] (1 species) 
Species Clostridium thermocellum [TaxId:1515] [101522] (2 PDB entries)
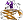
Chitin-binding protein CBP21 [117045] (1 species)
member of Pfam PF03067; elaborated fold with a large insertion between strands A and A'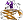
Species Serratia marcescens [TaxId:615] [117046] (3 PDB entries)
Uniprot O83009 28-197

Chitinase A, N-terminal domain N [49233] (1 species)
precedes the catalytic (beta/alpha)8-barrel domain
Species Serratia marcescens [TaxId:615] [49234] (11 PDB entries)
Uniprot P07254 24-563
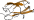
Cyclomaltodextrin glycanotransferase, domain D [49215] (4 species)
follows the starch-binding domain C; the catalytic domain A has (beta/alpha)8-barrel fold; family 13 glycosyl hydrolases
Species Bacillus circulans, different strains [TaxId:1397] [49216] (36 PDB entries) 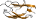
Species Bacillus sp., strain 1011 [TaxId:1409] [49219] (8 PDB entries)
Uniprot P05618
Species Bacillus stearothermophilus [TaxId:1422] [49217] (1 PDB entry) 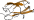
Species Thermoanaerobacterium thermosulfurigenes, EM1 [TaxId:33950] [49220] (4 PDB entries)

Cyclomaltodextrinase, N-terminal domain [101523] (1 species)
protein shares similar domain organization with maltogenic amylases but differs in the spatial arrangement of its domains
Species Flavobacterium sp. 92 [TaxId:197856] [101524] (1 PDB entry)

Five domain 'maltogenic' alpha-amylase (glucan 1,4-alpha-maltohydrolase), domain D [81280] (1 species)
domain architecture similar to cyclomaltodextrin glycosylhydrolases
Species Bacillus stearothermophilus [TaxId:1422] [81281] (2 PDB entries)

Galactose oxidase, C-terminal domain [49209] (3 species)
follows the catalytic seven-bladed beta-propeller domain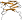
Species Dactylium dendroides [TaxId:5132] [49210] (3 PDB entries)
Uniprot Q01745 42-680
Species Fungus (Fusarium sp.) [TaxId:29916] [69164] (2 PDB entries)
sequence identical to that of Dactylium dendroides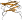
Species Fusarium graminearum (Gibberella zeae) [TaxId:5518] [158881] (6 PDB entries)

Glucans biosynthesis protein G (MdoG, OpgG), C-terminal domain [110054] (1 species) 
Species Escherichia coli [TaxId:562] [110055] (1 PDB entry)
Uniprot P33136 23-511

Glucodextranase, domain B [101525] (1 species) 
Species Arthrobacter globiformis [TaxId:1665] [101526] (2 PDB entries)
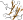
Glycosyltrehalose trehalohydrolase, N-terminal domain N [49224] (2 species)
domain architecture similar to isoamylase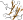
Species Deinococcus radiodurans [TaxId:1299] [141019] (9 PDB entries)
Uniprot Q9RX51 14-110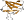
Species Sulfolobus solfataricus, km1 [TaxId:2287] [49225] (8 PDB entries)

Hyaluronate lyase precatalytic domain [69167] (1 species)
precedes the catalytic incomplete alpha5/alpha5 barrel
a rudiment form of Ig-like domain
Species Streptococcus agalactiae [TaxId:1311] [69168] (3 PDB entries)

Isoamylase, N-terminal domain N [49226] (1 species)
elaborated with a few large insertions in the common fold
precedes the catalytic (beta/alpha)8-barrel domain, the domain architecture similar to maltogenic amylases
Species Pseudomonas amyloderamosa [TaxId:32043] [49227] (1 PDB entry)
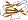
Maltogenic amylase, N-terminal domain N [49221] (4 species)
precedes the catalytic (beta/alpha)8-barrel domain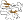
Species Bacillus sp., cyclomaltodextrinase [TaxId:1409] [74842] (1 PDB entry) 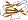
Species Thermoactinomyces vulgaris, TVAI [TaxId:2026] [74843] (8 PDB entries) 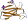
Species Thermoactinomyces vulgaris, TVAII [TaxId:2026] [49223] (12 PDB entries) 
Species Thermus sp. [TaxId:275] [49222] (2 PDB entries)

Neopullulanase, N-terminal domain [81960] (1 species)
homologous to maltogenic amylase
Species Bacillus stearothermophilus [TaxId:1422] [81961] (4 PDB entries)

Pullulanase PulA [158883] (1 species)
contains two E-set domains in tandem
Species Klebsiella pneumoniae [TaxId:573] [158884] (6 PDB entries)
Uniprot P07206 170-294! Uniprot P07206 295-409

Sialidase, linker domain [49237] (1 species)
follows the catalytic six-bladed beta-propeller domain
Species Micromonospora viridifaciens [TaxId:1881] [49238] (4 PDB entries)
Uniprot Q02834 47-647
More info for Family b.1.18.2: E-set domains of sugar-utilizing enzymes
Timeline for Family b.1.18.2: E-set domains of sugar-utilizing enzymes:
- Family b.1.18.2: E-set domains of sugar-utilizing enzymes first appeared in SCOP 1.63
- Family b.1.18.2: E-set domains of sugar-utilizing enzymes appears in SCOPe 2.07