Lineage for d1kxjb1 (1kxj B:1-199)
- Root: SCOPe 2.08

Class c: Alpha and beta proteins (a/b) [51349] (148 folds) 
Fold c.23: Flavodoxin-like [52171] (15 superfamilies)
3 layers, a/b/a; parallel beta-sheet of 5 strand, order 21345
Superfamily c.23.16: Class I glutamine amidotransferase-like [52317] (10 families) 
conserved positions of the oxyanion hole and catalytic nucleophile; different constituent families contain different additional structures
Family c.23.16.1: Class I glutamine amidotransferases (GAT) [52318] (12 proteins)
contains a catalytic Cys-His-Glu triad
Protein GAT subunit, HisH, (or domain) of imidazoleglycerolphosphate synthase HisF [69447] (4 species) 
Species Thermotoga maritima [TaxId:2336] [69448] (9 PDB entries) 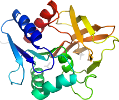
Domain d1kxjb1: 1kxj B:1-199 [73154]
Other proteins in same PDB: d1kxja2, d1kxjb2
complexed with po4
Details for d1kxjb1
PDB Entry: 1kxj (more details), 2.8 Å
PDB Description: the crystal structure of glutamine amidotransferase from thermotoga maritima
PDB Compounds: (B:) amidotransferase hishSCOPe Domain Sequences for d1kxjb1:
Sequence; same for both SEQRES and ATOM records: (download)
>d1kxjb1 c.23.16.1 (B:1-199) GAT subunit, HisH, (or domain) of imidazoleglycerolphosphate synthase HisF {Thermotoga maritima [TaxId: 2336]}
mrigiisvgpgnimnlyrgvkrasenfedvsielvesprndlydllfipgvghfgegmrr
lrendlidfvrkhvederyvvgvclgmqllfeeseeapgvkglsliegnvvklrsrrlph
mgwnevifkdtfpngyyyfvhtyravceeehvlgtteydgeifpsavrkgrilgfqfhpe
ksskigrkllekviecsls
SCOPe Domain Coordinates for d1kxjb1:
Click to download the PDB-style file with coordinates for d1kxjb1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1kxjb1:
- d1kxjb1 first appeared in SCOP 1.61, called d1kxjb_
- d1kxjb1 appears in SCOPe 2.07
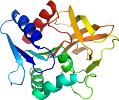
 View in 3D
View in 3D