Lineage for d3c9fb2 (3c9f B:16-337)
- Root: SCOPe 2.08

Class d: Alpha and beta proteins (a+b) [53931] (396 folds) 
Fold d.159: Metallo-dependent phosphatases [56299] (1 superfamily)
4 layers: alpha/beta/beta/alpha; mixed beta sheets; contains duplication
Superfamily d.159.1: Metallo-dependent phosphatases [56300] (13 families) 
different families of this superfamily are groupped in a single Pfam family, Pfam PF00149
Family d.159.1.2: 5'-nucleotidase (syn. UDP-sugar hydrolase), N-terminal domain [56307] (2 proteins) 
Protein 5'-nucleotidase (syn. UDP-sugar hydrolase), N-terminal domain [56308] (3 species) 
Species Yeast (Candida albicans) [TaxId:5476] [160868] (1 PDB entry)
Uniprot Q5A5Q7 16-337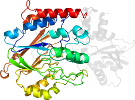
Domain d3c9fb2: 3c9f B:16-337 [156128]
Other proteins in same PDB: d3c9fa1, d3c9fa3, d3c9fb1
automated match to d3c9fa2
complexed with fmt, na, zn
Details for d3c9fb2
PDB Entry: 3c9f (more details), 1.9 Å
PDB Description: crystal structure of 5'-nucleotidase from candida albicans sc5314
PDB Compounds: (B:) 5'-nucleotidaseSCOPe Domain Sequences for d3c9fb2:
Sequence; same for both SEQRES and ATOM records: (download)
>d3c9fb2 d.159.1.2 (B:16-337) 5'-nucleotidase (syn. UDP-sugar hydrolase), N-terminal domain {Yeast (Candida albicans) [TaxId: 5476]}
sfphrnltwndinfvhttdthgwysghinqplyhanwgdfisftthmrriahsrnqdlll
idsgdrhdgnglsditspnglkstpifikqdydlltignhelylwenskqeyetvvnhfq
dkyvcsnvdirldnglfvplglkykyfttpirgirvmafgflfdfkrfnsgtrvtpmaet
ihepwfqealkhevdliiivghtpishnwgefyqvhqylrqffpdtiiqyfgghshirdf
tvfdslstglqsgrycetvgwtsvnldkadlnlpvrqrfsrsyidfntdsfkyhtnldke
fdtakgklvskliretrkelkl
SCOPe Domain Coordinates for d3c9fb2:
Click to download the PDB-style file with coordinates for d3c9fb2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d3c9fb2:
- d3c9fb2 first appeared in SCOP 1.75
- d3c9fb2 appears in SCOPe 2.07
 View in 3D
View in 3D