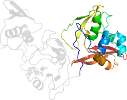
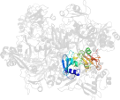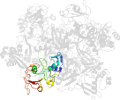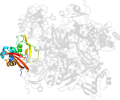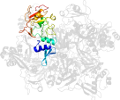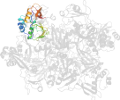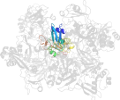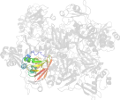Lineage for d2rhbe1 (2rhb E:1-190)
- Root: SCOPe 2.07

Class c: Alpha and beta proteins (a/b) [51349] (148 folds) 
Fold c.66: S-adenosyl-L-methionine-dependent methyltransferases [53334] (1 superfamily)
core: 3 layers, a/b/a; mixed beta-sheet of 7 strands, order 3214576; strand 7 is antiparallel to the rest
Superfamily c.66.1: S-adenosyl-L-methionine-dependent methyltransferases [53335] (60 families) 

Family c.66.1.48: Nsp15 N-terminal domain-like [142625] (2 proteins)
rudiment methyltransferase fold that probably has lost the enzymatic activity; contains extra N-terminal alpha+beta subdomain
Protein Nsp15 [142626] (2 species) 
Species SARS coronavirus [TaxId:227859] [142628] (3 PDB entries)
Uniprot Q6VA80 6619-6774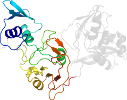
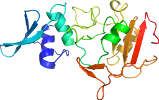
Domain d2rhbe1: 2rhb E:1-190 [152044]
Other proteins in same PDB: d2rhba2, d2rhba3, d2rhbb2, d2rhbb3, d2rhbc2, d2rhbc3, d2rhbd2, d2rhbd3, d2rhbe2, d2rhbe3, d2rhbf2
automated match to d2h85a1
mutant
Details for d2rhbe1
PDB Entry: 2rhb (more details), 2.8 Å
PDB Description: crystal structure of nsp15-h234a mutant- hexamer in asymmetric unit
PDB Compounds: (E:) Uridylate-specific endoribonucleaseSCOPe Domain Sequences for d2rhbe1:
Sequence; same for both SEQRES and ATOM records: (download)
>d2rhbe1 c.66.1.48 (E:1-190) Nsp15 {SARS coronavirus [TaxId: 227859]}
slenvaynvvnkghfdghageapvsiinnavytkvdgidveifenkttlpvnvafelwak
rnikpvpeikilnnlgvdiaantviwdykreapahvstigvctmtdiakkptesacsslt
vlfdgrvegqvdlfrnarngvlitegsvkgltpskgpaqasvngvtligesvktqfnyfk
kvdgiiqqlp
SCOPe Domain Coordinates for d2rhbe1:
Click to download the PDB-style file with coordinates for d2rhbe1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d2rhbe1:
- d2rhbe1 first appeared in SCOP 1.75
- d2rhbe1 appears in SCOPe 2.06
- d2rhbe1 appears in the current release, SCOPe 2.08
 View in 3D
View in 3D