Lineage for d1ezc_2 (1ezc 1-21,145-259)
- Root: SCOP 1.63

Class c: Alpha and beta proteins (a/b) [51349] (117 folds) 
Fold c.8: The "swivelling" beta/beta/alpha domain [52008] (6 superfamilies)
3 layers: b/b/a; the central sheet is parallel, and the other one is antiparallel; there are some variations in topology
this domain is thought to be mobile in all proteins known to contain it
Superfamily c.8.1: Phosphohistidine domain [52009] (2 families) 
contains barrel, closed, n=7, S=10
Family c.8.1.2: N-terminal domain of enzyme I of the PEP:sugar phosphotransferase system [52013] (1 protein) 
Protein N-terminal domain of enzyme I of the PEP:sugar phosphotransferase system [52014] (1 species)
contains 4-helical insert domain, residues 22-144
Species Escherichia coli [TaxId:562] [52015] (11 PDB entries) 
Domain d1ezc_2: 1ezc 1-21,145-259 [30713]
Other proteins in same PDB: d1ezc_1
Details for d1ezc_2
PDB Entry: 1ezc (more details)
PDB Description: amino terminal domain of enzyme i from escherichia coli, nmr, 17 structures
SCOP Domain Sequences for d1ezc_2:
Sequence; same for both SEQRES and ATOM records: (download)
>d1ezc_2 c.8.1.2 (1-21,145-259) N-terminal domain of enzyme I of the PEP:sugar phosphotransferase system {Escherichia coli}
misgilaspgiafgkalllkeXkiidlsaiqdevilvaadltpsetaqlnlkkvlgfitd
aggrtshtsimarslelpaivgtgsvtsqvknddylildavnnqvyvnptnevidkmrav
qeqvasekaelaklkdr
SCOP Domain Coordinates for d1ezc_2:
Click to download the PDB-style file with coordinates for d1ezc_2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1ezc_2:
- d1ezc_2 first appeared (with stable ids) in SCOP 1.55
- d1ezc_2 appears in SCOP 1.61
- d1ezc_2 appears in SCOP 1.65
- d1ezc_2 appears in the current release, SCOPe 2.08, called d1ezca2
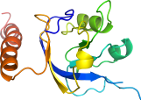
 View in 3D
View in 3D