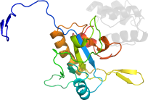
Lineage for d2bgxa4 (2bgx A:180-260)
- Root: SCOPe 2.08

Class a: All alpha proteins [46456] (290 folds) 
Fold a.20: PGBD-like [47089] (1 superfamily)
core: 3 helices; bundle, closed, left-handed twist; parallel
Superfamily a.20.1: PGBD-like [47090] (3 families) 

Family a.20.1.1: Peptidoglycan binding domain, PGBD [47091] (2 proteins) 
Protein Probable N-acetylmuramoyl-L-alanine amidase YbjR, C-terminal domain [140390] (1 species) 
Species Escherichia coli [TaxId:562] [140391] (3 PDB entries)
Uniprot P75820 195-275
Domain d2bgxa4: 2bgx A:180-260 [303580]
Other proteins in same PDB: d2bgxa3
automated match to d2wkxa1
complexed with zn
Details for d2bgxa4
PDB Entry: 2bgx (more details), 1.8 Å
PDB Description: crystal structure of native amid at 1.8 angstrom
PDB Compounds: (A:) n-acetylmuramoyl-l-alanine amidaseSCOPe Domain Sequences for d2bgxa4:
Sequence; same for both SEQRES and ATOM records: (download)
>d2bgxa4 a.20.1.1 (A:180-260) Probable N-acetylmuramoyl-L-alanine amidase YbjR, C-terminal domain {Escherichia coli [TaxId: 562]}
pdaqrvnfylagraphtpvdtasllellarygydvkpdmtpreqrrvimafqmhfrptly
ngeadaetqaiaeallekygq
SCOPe Domain Coordinates for d2bgxa4:
Click to download the PDB-style file with coordinates for d2bgxa4.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d2bgxa4:
- d2bgxa4 first appeared in SCOPe 2.06
- d2bgxa4 appears in SCOPe 2.07
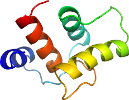
 View in 3D
View in 3D