Lineage for d1hv9b1 (1hv9 B:252-452)
- Root: SCOP 1.69

Class b: All beta proteins [48724] (144 folds) 
Fold b.81: Single-stranded left-handed beta-helix [51160] (3 superfamilies)
superhelix turns are made of parallel beta-strands and (short) turns
Superfamily b.81.1: Trimeric LpxA-like enzymes [51161] (6 families) 
superhelical turns are made of three short strands; duplication: the sequence hexapeptide repeats correspond to individual strands
Family b.81.1.4: N-acetylglucosamine 1-phosphate uridyltransferase GlmU, C-terminal domain [51171] (1 protein)
this is a repeat family; one repeat unit is 2oi5 A:321-339 found in domain
Protein N-acetylglucosamine 1-phosphate uridyltransferase GlmU, C-terminal domain [51172] (2 species) 
Species Escherichia coli [TaxId:562] [51173] (3 PDB entries) 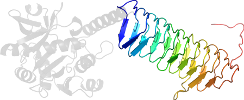
Domain d1hv9b1: 1hv9 B:252-452 [28061]
Other proteins in same PDB: d1hv9a2, d1hv9b2
Details for d1hv9b1
PDB Entry: 1hv9 (more details), 2.1 Å
PDB Description: structure of e. coli glmu: analysis of pyrophosphorylase and acetyltransferase active sites
SCOP Domain Sequences for d1hv9b1:
Sequence; same for both SEQRES and ATOM records: (download)
>d1hv9b1 b.81.1.4 (B:252-452) N-acetylglucosamine 1-phosphate uridyltransferase GlmU, C-terminal domain {Escherichia coli}
vmlrdparfdlrgtlthgrdveidtnviiegnvtlghrvkigtgcviknsvigddceisp
ytvvedanlaaactigpfarlrpgaellegahvgnfvemkkarlgkgskaghltylgdae
igdnvnigagtitcnydgankfktiigddvfvgsdtqlvapvtvgkgatiaagttvtrnv
genalaisrvpqtqkegwrrp
SCOP Domain Coordinates for d1hv9b1:
Click to download the PDB-style file with coordinates for d1hv9b1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1hv9b1:
- d1hv9b1 first appeared (with stable ids) in SCOP 1.55
- d1hv9b1 appears in SCOP 1.67
- d1hv9b1 appears in SCOP 1.71
- d1hv9b1 appears in the current release, SCOPe 2.08

 View in 3D
View in 3D