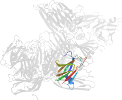
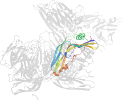
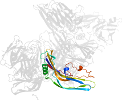
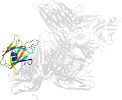
Lineage for d3ziwb2 (3ziw B:200-319)
- Root: SCOPe 2.07

Class b: All beta proteins [48724] (178 folds) 
Fold b.18: Galactose-binding domain-like [49784] (1 superfamily)
sandwich; 9 strands in 2 sheets; jelly-roll
Superfamily b.18.1: Galactose-binding domain-like [49785] (35 families) 

Family b.18.1.37: Clostridium perfringens enterotoxin (CPE), C-terminal domain [267611] (2 proteins)
C-terminal part of Pfam PF03505; PubMed 17977833 describes homology between (d2quoa_), (d1w99a3), and (d1nqdb_)
Protein automated matches [267681] (1 species)
not a true protein
Species Clostridium perfringens [TaxId:1502] [267916] (2 PDB entries) 
Domain d3ziwb2: 3ziw B:200-319 [265792]
Other proteins in same PDB: d3ziwa1, d3ziwa3, d3ziwb1, d3ziwb3, d3ziwc1, d3ziwc3, d3ziwd1, d3ziwd3, d3ziwe1, d3ziwe3, d3ziwf1, d3ziwf3
automated match to d3am2a1
complexed with p6g; mutant
Details for d3ziwb2
PDB Entry: 3ziw (more details), 1.9 Å
PDB Description: Clostridium perfringens enterotoxin, D48A mutation and N-terminal 37 residues deleted
PDB Compounds: (B:) heat-labile enterotoxin b chainSCOPe Domain Sequences for d3ziwb2:
Sequence; same for both SEQRES and ATOM records: (download)
>d3ziwb2 b.18.1.37 (B:200-319) automated matches {Clostridium perfringens [TaxId: 1502]}
ldlaaaterlnltdalnsnpagnlydwrssnsypwtqklnlhltitatgqkyrilaskiv
dfniysnnfnnlvkleqslgdgvkdhyvdisldagqyvlvmkanssysgnypysilfqkf
SCOPe Domain Coordinates for d3ziwb2:
Click to download the PDB-style file with coordinates for d3ziwb2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d3ziwb2:
- d3ziwb2 first appeared in SCOPe 2.05
- d3ziwb2 appears in SCOPe 2.06
- d3ziwb2 appears in the current release, SCOPe 2.08
 View in 3D
View in 3D