Lineage for d1ehga_ (1ehg A:)
- Root: SCOPe 2.08

Class a: All alpha proteins [46456] (290 folds) 
Fold a.104: Cytochrome P450 [48263] (1 superfamily)
multihelical
Superfamily a.104.1: Cytochrome P450 [48264] (2 families) 

Family a.104.1.1: Cytochrome P450 [48265] (23 proteins) 
Protein Cytochrome P450-NOR, nitric reductase [48270] (1 species) 
Species Fungus (Fusarium oxysporum) [TaxId:5507] [48271] (27 PDB entries)
Uniprot P23295
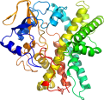
Domain d1ehga_: 1ehg A: [18956]
complexed with hem; mutant
Details for d1ehga_
PDB Entry: 1ehg (more details), 1.7 Å
PDB Description: crystal structures of cytochrome p450nor and its mutants (ser286 val, thr) in the ferric resting state at cryogenic temperature: a comparative analysis with monooxygenase cytochrome p450s
PDB Compounds: (A:) cytochrome p450norSCOPe Domain Sequences for d1ehga_:
Sequence; same for both SEQRES and ATOM records: (download)
>d1ehga_ a.104.1.1 (A:) Cytochrome P450-NOR, nitric reductase {Fungus (Fusarium oxysporum) [TaxId: 5507]}
apsfpfsrasgpeppaefaklratnpvsqvklfdgslawlvtkhkdvcfvatseklskvr
trqgfpelsasgkqaakakptfvdmdppehmhqrsmveptftpeavknlqpyiqrtvddl
leqmkqkgcangpvdlvkefalpvpsyiiytllgvpfndleyltqqnairtngsstarea
saanqelldylailveqrlvepkddiisklcteqvkpgnidksdavqiaflllvagnatm
vnmialgvatlaqhpdqlaqlkanpslapqfveelcryhtavalaikrtakedvmigdkl
vranegiiasnqsanrdeevfenpdefnmnrkwppqdplgfgfgdhrciaehlakaeltt
vfstlyqkfpdlkvavplgkinytplnrdvgivdlpvif
SCOPe Domain Coordinates for d1ehga_:
Click to download the PDB-style file with coordinates for d1ehga_.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1ehga_:
- d1ehga_ first appeared (with stable ids) in SCOP 1.55
- d1ehga_ appears in SCOPe 2.07
 View in 3D
View in 3D