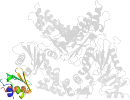Lineage for d3bp8d1 (3bp8 D:13-87)
- Root: SCOP 1.75

Class d: Alpha and beta proteins (a+b) [53931] (376 folds) 
Fold d.95: Homing endonuclease-like [55603] (2 superfamilies)
alpha-beta(2)-alpha-beta(2)-alpha; 2 layers: a/b; antiparallel sheet 1243
Superfamily d.95.1: Glucose permease domain IIB [55604] (1 family) 
Family d.95.1.1: Glucose permease domain IIB [55605] (1 protein) 
Protein Glucose permease domain IIB [55606] (1 species) 
Species Escherichia coli [TaxId:562] [55607] (3 PDB entries)
there are differences in secondary structure packing between the two NMR-determined structures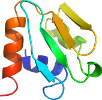
Domain d3bp8d1: 3bp8 D:13-87 [155470]
Other proteins in same PDB: d3bp8a1, d3bp8a2, d3bp8a3, d3bp8b1, d3bp8b2, d3bp8b3
automatically matched to d1ibaa_
complexed with act, zn
Details for d3bp8d1
PDB Entry: 3bp8 (more details), 2.85 Å
PDB Description: Crystal structure of Mlc/EIIB complex
PDB Compounds: (D:) PTS system glucose-specific EIICB componentSCOP Domain Sequences for d3bp8d1:
Sequence; same for both SEQRES and ATOM records: (download)
>d3bp8d1 d.95.1.1 (D:13-87) Glucose permease domain IIB {Escherichia coli [TaxId: 562]}
mapalvaafggkenitnldacitrlrvsvadvskvdqaglkklgaagvvvagsgvqaifg
tksdnlktemdeyir
SCOP Domain Coordinates for d3bp8d1:
Click to download the PDB-style file with coordinates for d3bp8d1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d3bp8d1:
- d3bp8d1 is new in SCOP 1.75
- d3bp8d1 is called d3bp8d_ in SCOPe 2.01
- d3bp8d1 appears in the current release, SCOPe 2.08, called d3bp8d_
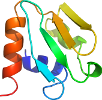
 View in 3D
View in 3D