Lineage for d2bvma_ (2bvm A:)
- Root: SCOPe 2.04

Class c: Alpha and beta proteins (a/b) [51349] (148 folds) 
Fold c.68: Nucleotide-diphospho-sugar transferases [53447] (1 superfamily)
3 layers: a/b/a; mixed beta-sheet of 7 strands, order 3214657; strand 6 is antiparallel to the rest
Superfamily c.68.1: Nucleotide-diphospho-sugar transferases [53448] (20 families) 

Family c.68.1.22: Glycosylating toxin catalytic domain-like [159726] (4 proteins)
contains Glycosyltransferase sugar-binding region containing DXD motif, Pfam PF04488
Protein automated matches [190532] (2 species)
not a true protein
Species Clostridium difficile [TaxId:1496] [187493] (1 PDB entry) 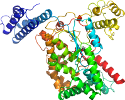
Domain d2bvma_: 2bvm A: [146220]
automated match to d2bvla1
complexed with glc, mn, so4, udp
Details for d2bvma_
PDB Entry: 2bvm (more details), 2.55 Å
PDB Description: crystal structure of the catalytic domain of toxin b from clostridium difficile in complex with udp, glc and manganese ion
PDB Compounds: (A:) toxin bSCOPe Domain Sequences for d2bvma_:
Sequence; same for both SEQRES and ATOM records: (download)
>d2bvma_ c.68.1.22 (A:) automated matches {Clostridium difficile [TaxId: 1496]}
mslvnrkqlekmanvrfrtqedeyvaildaleeyhnmsentvvekylklkdinsltdici
dtykksgrnkalkkfkeylvtevlelknnnltpveknlhfvwiggqindtainyinqwkd
vnsdynvnvfydsnaflintlkktvvesaindtlesfrenlndprfdynkffrkrmeiiy
dkqknfinyykaqreenpeliiddivktylsneyskeidelntyieeslnkitqnsgndv
rnfgefkngesfnlyeqelverwnlaaasdilrisalkeiggmyldvdmlpgiqpdlfes
iekpssvtvdfwemtkleaimkykeyipeytsehfdmldeevqssfesvlasksdkseif
sslgdmeasplevkiafnskgiinqglisvkdsycsnlivkqienrykilnnslnpaise
dndfntttntfidsimaeanadngrfmmelgkylrvgffpdvkttinlsgpeayaaayqd
llmfkegsmnihlieadlrnfeisktnisqsteqemaslwsfddarakaqfeeykrnyfe
ga
SCOPe Domain Coordinates for d2bvma_:
Click to download the PDB-style file with coordinates for d2bvma_.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d2bvma_:
- d2bvma_ first appeared in SCOP 1.75, called d2bvma1
- d2bvma_ appears in SCOPe 2.03
- d2bvma_ appears in SCOPe 2.05
- d2bvma_ appears in the current release, SCOPe 2.08
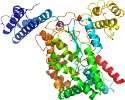
 View in 3D
View in 3D