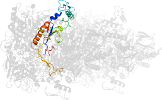
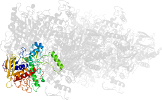
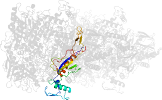Lineage for d1tzla1 (1tzl A:43-354,A:553-619)
- Root: SCOPe 2.07

Class c: Alpha and beta proteins (a/b) [51349] (148 folds) 
Fold c.3: FAD/NAD(P)-binding domain [51904] (1 superfamily)
core: 3 layers, b/b/a; central parallel beta-sheet of 5 strands, order 32145; top antiparallel beta-sheet of 3 strands, meander
Superfamily c.3.1: FAD/NAD(P)-binding domain [51905] (9 families) 

Family c.3.1.2: FAD-linked reductases, N-terminal domain [51913] (18 proteins)
C-terminal domain is alpha+beta is common for the family
Protein Pyranose 2-oxidase [117439] (1 species) 
Species White-rot fungus (Peniophora sp. SG) [TaxId:204723] [117440] (5 PDB entries)
Uniprot Q8J136 43-619
Domain d1tzla1: 1tzl A:43-354,A:553-619 [112876]
Other proteins in same PDB: d1tzla2, d1tzlb2, d1tzlc2, d1tzld2, d1tzle2, d1tzlf2, d1tzlg2, d1tzlh2
complexed with fad
Details for d1tzla1
PDB Entry: 1tzl (more details), 2.35 Å
PDB Description: crystal structure of pyranose 2-oxidase from the white-rot fungus peniophora sp.
PDB Compounds: (A:) Pyranose oxidaseSCOPe Domain Sequences for d1tzla1:
Sequence; same for both SEQRES and ATOM records: (download)
>d1tzla1 c.3.1.2 (A:43-354,A:553-619) Pyranose 2-oxidase {White-rot fungus (Peniophora sp. SG) [TaxId: 204723]}
mdikydvvivgsgpigctyarelvgagykvamfdigeidsglkigahkkntveyqknidk
fvnviqgqlmsvsvpvntlvvdtlsptswqastffvrngsnpeqdplrnlsgqavtrvvg
gmsthwtcatprfdreqrpllvkddadaddaewdrlytkaesyfqtgtdqfkesirhnlv
lnklaeeykgqrdfqqiplaatrrsptfvewssantvfdlqnrpntdapeerfnlfpava
cervvrnalnseieslhihdlisgdrfeikadvyvltagavhntqllvnsgfgqlgrpnp
tnppellpslgsXhrmgfdekednccvntdsrvfgfknlflggcgniptayganptltam
slaiksceyikqnftpspft
SCOPe Domain Coordinates for d1tzla1:
Click to download the PDB-style file with coordinates for d1tzla1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1tzla1:
- d1tzla1 first appeared in SCOP 1.71
- d1tzla1 appears in SCOPe 2.06
- d1tzla1 appears in the current release, SCOPe 2.08

 View in 3D
View in 3D