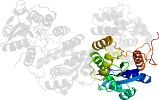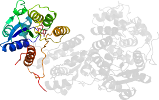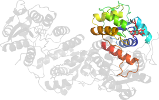Lineage for d1sbzd_ (1sbz D:)
- Root: SCOPe 2.01

Class c: Alpha and beta proteins (a/b) [51349] (147 folds) 
Fold c.34: Homo-oligomeric flavin-containing Cys decarboxylases, HFCD [52506] (1 superfamily)
3 layers: a/b/a; parallel beta-sheet of 6 strands, order 321456
Superfamily c.34.1: Homo-oligomeric flavin-containing Cys decarboxylases, HFCD [52507] (2 families) 
Family c.34.1.1: Homo-oligomeric flavin-containing Cys decarboxylases, HFCD [52508] (4 proteins)
Pfam PF02441; formerly DFP, DNA/pantothenate metabolism flavoprotein family
Protein Probable aromatic acid decarboxylase Pad1 [117500] (1 species) 
Species Escherichia coli O157:H7 [TaxId:83334] [117501] (1 PDB entry)
Uniprot Q9X728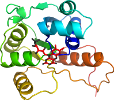
Domain d1sbzd_: 1sbz D: [112071]
complexed with fmn
Details for d1sbzd_
PDB Entry: 1sbz (more details), 2 Å
PDB Description: crystal structure of dodecameric fmn-dependent ubix-like decarboxylase from escherichia coli o157:h7
PDB Compounds: (D:) Probable aromatic acid decarboxylaseSCOPe Domain Sequences for d1sbzd_:
Sequence; same for both SEQRES and ATOM records: (download)
>d1sbzd_ c.34.1.1 (D:) Probable aromatic acid decarboxylase Pad1 {Escherichia coli O157:H7 [TaxId: 83334]}
mklivgmtgatgaplgvallqalrempnvethlvmskwakttieletpysardvaaladf
shnpadqaatissgsfrtdgmivipcsmktlagiragyadglvgraadvvlkegrklvlv
premplstihlenmlalsrmgvamvppmpafynhpetvddivhhvvarvldqfglehpya
rrwqg
SCOPe Domain Coordinates for d1sbzd_:
Click to download the PDB-style file with coordinates for d1sbzd_.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1sbzd_:
- d1sbzd_ first appeared in SCOP 1.71
- d1sbzd_ appears in SCOP 1.75
- d1sbzd_ appears in SCOPe 2.02
- d1sbzd_ appears in the current release, SCOPe 2.08
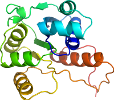
 View in 3D
View in 3D