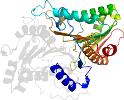
Lineage for d1v0ua2 (1v0u A:264-513)
- Root: SCOPe 2.06

Class d: Alpha and beta proteins (a+b) [53931] (385 folds) 
Fold d.136: Phospholipase D/nuclease [56023] (1 superfamily)
beta-alpha-beta-alpha-beta-alpha-beta(4)-alpha; mixed sheet: order 1765234
Superfamily d.136.1: Phospholipase D/nuclease [56024] (6 families) 

Family d.136.1.2: Phospholipase D [64391] (1 protein)
duplication: contains two domains of this fold arranged as in the Nuc dimer
Protein Phospholipase D [64392] (1 species) 
Species Streptomyces sp. [TaxId:1931] [64393] (8 PDB entries)
Uniprot P84147
Domain d1v0ua2: 1v0u A:264-513 [108217]
complexed with po3
Details for d1v0ua2
PDB Entry: 1v0u (more details), 1.42 Å
PDB Description: phospholipase d from streptomyces sp. strain pmf soaked with the product glycerophosphate.
PDB Compounds: (A:) phospholipase dSCOPe Domain Sequences for d1v0ua2:
Sequence, based on SEQRES records: (download)
>d1v0ua2 d.136.1.2 (A:264-513) Phospholipase D {Streptomyces sp. [TaxId: 1931]}
nvpviavgglgvgikdvdpkstfrpdlptasdtkcvvglhdntnadrdydtvnpeesalr
alvasakghieisqqdlnatcpplprydirlydalaakmaagvkvrivvsdpanrgavgs
ggysqikslseisdtlrnrlanitggqqaaktamcsnlqlatfrsspngkwadghpyaqh
hklvsvdsstfyigsknlypswlqdfgyivespeaakqldaklldpqwkysqetatvdya
rgicn
Sequence, based on observed residues (ATOM records): (download)
>d1v0ua2 d.136.1.2 (A:264-513) Phospholipase D {Streptomyces sp. [TaxId: 1931]}
nvpviavgglgvgikdvdpkstfrpdlptasdtkcvvglhdntnadrdydtvnpeesalr
alvasakghieisqqdlnatcpplprydirlydalaakmaagvkvrivvsdpanrgysqi
kslseisdtlrnrlanitggqqaaktamcsnlqlatfrsspngkwadghpyaqhhklvsv
dsstfyigsknlypswlqdfgyivespeaakqldaklldpqwkysqetatvdyargicn
SCOPe Domain Coordinates for d1v0ua2:
Click to download the PDB-style file with coordinates for d1v0ua2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1v0ua2:
- d1v0ua2 first appeared in SCOP 1.69
- d1v0ua2 appears in SCOPe 2.05
- d1v0ua2 appears in SCOPe 2.07
- d1v0ua2 appears in the current release, SCOPe 2.08
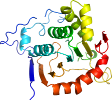
 View in 3D
View in 3D