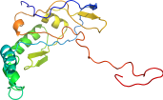
Lineage for d1t1ea2 (1t1e A:12-190)
- Root: SCOPe 2.01

Class d: Alpha and beta proteins (a+b) [53931] (376 folds) 
Fold d.58: Ferredoxin-like [54861] (59 superfamilies)
alpha+beta sandwich with antiparallel beta-sheet; (beta-alpha-beta)x2
Superfamily d.58.3: Protease propeptides/inhibitors [54897] (3 families) 

Family d.58.3.2: Subtilase propeptides/inhibitors [54905] (4 proteins)
decorated with additional structure
Protein Pro-kumamolisin activation domain [110951] (1 species) 
Species Bacillus sp. MN-32 [TaxId:198803] [110952] (1 PDB entry)
Uniprot Q8RR56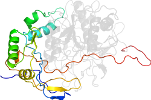
Domain d1t1ea2: 1t1e A:12-190 [106245]
Other proteins in same PDB: d1t1ea1
complexed with ca
Details for d1t1ea2
PDB Entry: 1t1e (more details), 1.18 Å
PDB Description: high resolution crystal structure of the intact pro-kumamolisin, a sedolisin type proteinase (previously called kumamolysin or kscp)
PDB Compounds: (A:) kumamolisinSCOPe Domain Sequences for d1t1ea2:
Sequence; same for both SEQRES and ATOM records: (download)
>d1t1ea2 d.58.3.2 (A:12-190) Pro-kumamolisin activation domain {Bacillus sp. MN-32 [TaxId: 198803]}
ekrevlagharrqapqavdkgpvtgdqrisvtvvlrrqrgdeleahverqaalapharvh
lereafaashgaslddfaeirkfaeahgltldrahvaagtavlsgpvdavnqafgvelrh
fdhpdgsyrsyvgdvrvpasiaplieavlgldtrpvarphfrlrrraegefearsqsaa
SCOPe Domain Coordinates for d1t1ea2:
Click to download the PDB-style file with coordinates for d1t1ea2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1t1ea2:
- d1t1ea2 first appeared in SCOP 1.69
- d1t1ea2 appears in SCOP 1.75
- d1t1ea2 appears in SCOPe 2.02
- d1t1ea2 appears in the current release, SCOPe 2.08
 View in 3D
View in 3D