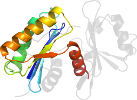
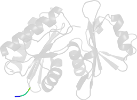
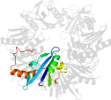
Lineage for d1okjd2 (1okj D:107-216)
- Root: SCOPe 2.08

Class c: Alpha and beta proteins (a/b) [51349] (148 folds) 
Fold c.55: Ribonuclease H-like motif [53066] (7 superfamilies)
3 layers: a/b/a; mixed beta-sheet of 5 strands, order 32145; strand 2 is antiparallel to the rest
Superfamily c.55.1: Actin-like ATPase domain [53067] (16 families) 
duplication contains two domains of this fold
Family c.55.1.9: YeaZ-like [110633] (3 proteins)
Pfam PF00814; ubiquitous cytoplasmic protein; annotated as Glycoprotease (Peptidase_M22 family) on the basis of one member's known extracellular activity
Protein Hypothetical protein YeaZ [110634] (1 species) 
Species Escherichia coli [TaxId:562] [110635] (1 PDB entry)
Uniprot P76256
Domain d1okjd2: 1okj D:107-216 [104005]
Other proteins in same PDB: d1okja3, d1okjb3, d1okjc3, d1okjd3
complexed with gd3
Details for d1okjd2
PDB Entry: 1okj (more details), 2.28 Å
PDB Description: crystal structure of the essential E. coli YeaZ protein by MAD method using the gadolinium complex "DOTMA"
PDB Compounds: (D:) tRNA threonylcarbamoyladenosine biosynthesis protein tsabSCOPe Domain Sequences for d1okjd2:
Sequence; same for both SEQRES and ATOM records: (download)
>d1okjd2 c.55.1.9 (D:107-216) Hypothetical protein YeaZ {Escherichia coli [TaxId: 562]}
atrvlaaidarmgevywaeyqrdengiwhgeeteavlkpeivhermqqlsgewvtvgtgw
qawpdlgkesglvlrdgevllpaaedmlpiacqmfaegktvavehaepvy
SCOPe Domain Coordinates for d1okjd2:
Click to download the PDB-style file with coordinates for d1okjd2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1okjd2:
- d1okjd2 first appeared in SCOP 1.69
- d1okjd2 appears in SCOPe 2.07

 View in 3D
View in 3D