Lineage for d4ixna1 (4ixn A:1-223)
- Root: SCOPe 2.07

Class c: Alpha and beta proteins (a/b) [51349] (148 folds) 
Fold c.37: P-loop containing nucleoside triphosphate hydrolases [52539] (1 superfamily)
3 layers: a/b/a, parallel or mixed beta-sheets of variable sizes
Superfamily c.37.1: P-loop containing nucleoside triphosphate hydrolases [52540] (26 families) 
division into families based on beta-sheet topologies
Family c.37.1.10: Nitrogenase iron protein-like [52652] (16 proteins)
core: parallel beta-sheet of 7 strands; order 3241567
Protein Hypothetical protein YjiA, N-terminal domain [89671] (1 species)
homologue of the cobalamin biosynthesis protein CobW
Species Escherichia coli [TaxId:562] [89672] (3 PDB entries) 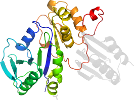
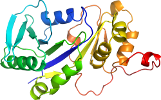
Domain d4ixna1: 4ixn A:1-223 [234918]
Other proteins in same PDB: d4ixna2, d4ixnb2
automated match to d1nija1
complexed with so4, zn; mutant
Details for d4ixna1
PDB Entry: 4ixn (more details), 2.05 Å
PDB Description: Crystal Structure of Zn(II)-bound E37A,C66A,C67A triple mutant YjiA GTPase
PDB Compounds: (A:) Uncharacterized GTP-binding protein YjiASCOPe Domain Sequences for d4ixna1:
Sequence; same for both SEQRES and ATOM records: (download)
>d4ixna1 c.37.1.10 (A:1-223) Hypothetical protein YjiA, N-terminal domain {Escherichia coli [TaxId: 562]}
mnpiavtlltgflgagkttllrhilneqhgykiavianefgevsvddqligdratqiktl
tngciaasrsneledalldlldnldkgniqfdrlviectgmadpgpiiqtffshevlcqr
ylldgvialvdavhadeqmnqftiaqsqvgyadrilltktdvageaeklherlarinara
pvytvthgdidlgllfntngfmleenvvstkprfhfiadkqnd
SCOPe Domain Coordinates for d4ixna1:
Click to download the PDB-style file with coordinates for d4ixna1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d4ixna1:
- d4ixna1 first appeared in SCOPe 2.03
- d4ixna1 appears in SCOPe 2.06
- d4ixna1 appears in the current release, SCOPe 2.08
 View in 3D
View in 3D