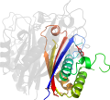
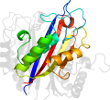
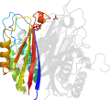Lineage for d3ikfa_ (3ikf A:)
- Root: SCOPe 2.08

Class d: Alpha and beta proteins (a+b) [53931] (396 folds) 
Fold d.79: Bacillus chorismate mutase-like [55297] (9 superfamilies)
core: beta-alpha-beta-alpha-beta(2); mixed beta-sheet: order: 1423, strand 4 is antiparallel to the rest
Superfamily d.79.5: IpsF-like [69765] (2 families) 
forms trimers with three closely packed beta-sheets; possible link between the YjgF-like (d.79.1) and 4'-phosphopantetheinyl transferase superfamilies (d.150.1)
Family d.79.5.1: IpsF-like [69766] (3 proteins)
automatically mapped to Pfam PF02542
Protein automated matches [190985] (6 species)
not a true protein
Species Burkholderia pseudomallei [TaxId:28450] [189021] (2 PDB entries) 
Domain d3ikfa_: 3ikf A: [178372]
automated match to d1jn1a_
complexed with 717, act, cl, k, zn
Details for d3ikfa_
PDB Entry: 3ikf (more details), 2.07 Å
PDB Description: Crystal structure of 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase from Burkholderia pseudomallei with FOL fragment 717, imidazo[2,,1-b][1,3]thiazol-6-ylmethanol
PDB Compounds: (A:) 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthaseSCOPe Domain Sequences for d3ikfa_:
Sequence; same for both SEQRES and ATOM records: (download)
>d3ikfa_ d.79.5.1 (A:) automated matches {Burkholderia pseudomallei [TaxId: 28450]}
mdfrigqgydvhqlvpgrpliiggvtipyergllghsdadvllhaitdalfgaaalgdig
rhfsdtdprfkgadsrallrecasrvaqagfairnvdstiiaqapklaphidamraniaa
dldlpldrvnvkaktneklgylgrgegieaqaaalvvr
SCOPe Domain Coordinates for d3ikfa_:
Click to download the PDB-style file with coordinates for d3ikfa_.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d3ikfa_:
- d3ikfa_ first appeared in SCOPe 2.01
- d3ikfa_ appears in SCOPe 2.07
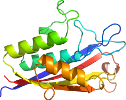
 View in 3D
View in 3D