Lineage for d7cufa2 (7cuf A:159-310)
- Root: SCOPe 2.07

Class d: Alpha and beta proteins (a+b) [53931] (388 folds) 
Fold d.96: T-fold [55619] (2 superfamilies)
beta(2)-alpha(2)-beta(2); 2 layers: alpha/beta; antiparallel sheet 1234
tunnel-shaped: its known members form wide oligomeric barrels different sizes
Superfamily d.96.1: Tetrahydrobiopterin biosynthesis enzymes-like [55620] (5 families) 
bind purine or pterin in topologically similar sites between subunits
Family d.96.1.4: Urate oxidase (uricase) [55633] (2 proteins)
automatically mapped to Pfam PF01014
Protein automated matches [254656] (4 species)
not a true protein
Species Bacillus sp. [TaxId:36824] [312567] (14 PDB entries) 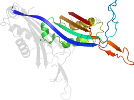
Domain d7cufa2: 7cuf A:159-310 [394514]
automated match to d1j2ga2
complexed with aza, mxe, na, oxy, so4
Details for d7cufa2
PDB Entry: 7cuf (more details), 1.46 Å
PDB Description: crystal structure of urate oxidase from bacillus sp. tb-90 in the absence from chloride anion at 1.44 a resolution
PDB Compounds: (A:) Uric acid degradation bifunctional proteinSCOPe Domain Sequences for d7cufa2:
Sequence; same for both SEQRES and ATOM records: (download)
>d7cufa2 d.96.1.4 (A:159-310) automated matches {Bacillus sp. [TaxId: 36824]}
dntlniteqqsglaglqlikvsgnsfvgfirdeyttlpedsnrplfvylnikwkyknted
sfgtnpenyvaaeqirdiatsvfhetetlsiqhliyligrrilerfpqlqevyfesqnht
wdkiveeipesegkvyteprppygfqcftvtq
SCOPe Domain Coordinates for d7cufa2:
Click to download the PDB-style file with coordinates for d7cufa2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d7cufa2:
- d7cufa2 is new in SCOPe 2.07-stable
- d7cufa2 appears in the current release, SCOPe 2.08

 View in 3D
View in 3D