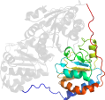
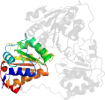
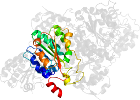
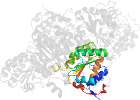
Lineage for d1pyda3 (1pyd A:361-556)
- Root: SCOP 1.55

Class c: Alpha and beta proteins (a/b) [51349] (97 folds) 
Fold c.36: Thiamin diphosphate-binding fold (THDP-binding) [52517] (1 superfamily) 
Superfamily c.36.1: Thiamin diphosphate-binding fold (THDP-binding) [52518] (4 families) 

Family c.36.1.1: Pyruvate oxidase and decarboxylase [52519] (3 proteins) 
Protein Pyruvate decarboxylase [52520] (3 species) 
Species Brewer's yeast (Saccharomyces), uvarum strain [TaxId:230603] [52521] (1 PDB entry) 
Domain d1pyda3: 1pyd A:361-556 [31774]
Other proteins in same PDB: d1pyda1, d1pydb1
Details for d1pyda3
PDB Entry: 1pyd (more details), 2.4 Å
PDB Description: catalytic centers in the thiamin diphosphate dependent enzyme pyruvate decarboxylase at 2.4 angstroms resolution
SCOP Domain Sequences for d1pyda3:
Sequence; same for both SEQRES and ATOM records: (download)
>d1pyda3 c.36.1.1 (A:361-556) Pyruvate decarboxylase {Brewer's yeast (Saccharomyces), uvarum strain}
astplkqewmwnqlgnflqegdvviaetgtsafginqttfpnntygisqvlwgsigfttg
atlgaafaaeeidpkkrvilfigdgslqltvqeistmirwglkpylfvlnndgytiekli
hgpkaqyneiqgwdhlsllptfgakdyethrvattgewdkltqdksfndnskirmieiml
pvfdapqnlvkqaklt
SCOP Domain Coordinates for d1pyda3:
Click to download the PDB-style file with coordinates for d1pyda3.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1pyda3:
- d1pyda3 appears in SCOP 1.57
- d1pyda3 appears in the current release, SCOPe 2.08
 View in 3D
View in 3D