Lineage for d3schb2 (3sch B:77-198)
- Root: SCOPe 2.03

Class b: All beta proteins [48724] (174 folds) 
Fold b.82: Double-stranded beta-helix [51181] (7 superfamilies)
one turn of helix is made by two pairs of antiparallel strands linked with short turns
has appearance of a sandwich of distinct architecture and jelly-roll topology
Superfamily b.82.1: RmlC-like cupins [51182] (25 families) 

Family b.82.1.10: TM1459-like [101976] (2 proteins) 
Protein Hydroxypropylphosphonic acid epoxidase Fom4, C-terminal domain [141593] (1 species) 
Species Streptomyces wedmorensis [TaxId:43759] [141594] (13 PDB entries)
Uniprot Q56185 77-198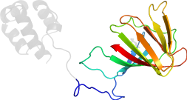
Domain d3schb2: 3sch B:77-198 [216324]
Other proteins in same PDB: d3scha1, d3schb1
automated match to d1zz6a2
complexed with co, tb6
Details for d3schb2
PDB Entry: 3sch (more details), 2.1 Å
PDB Description: co(ii)-hppe with r-hpp
PDB Compounds: (B:) epoxidaseSCOPe Domain Sequences for d3schb2:
Sequence; same for both SEQRES and ATOM records: (download)
>d3schb2 b.82.1.10 (B:77-198) Hydroxypropylphosphonic acid epoxidase Fom4, C-terminal domain {Streptomyces wedmorensis [TaxId: 43759]}
dlddgviiqmpderpilkgvrdnvdyyvynclvrtkrapslvplvvdvltdnpddakfns
ghagneflfvlegeihmkwgdkenpkeallptgasmfveehvphaftaakgtgsakliav
nf
SCOPe Domain Coordinates for d3schb2:
Click to download the PDB-style file with coordinates for d3schb2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d3schb2:
- d3schb2 is new in SCOPe 2.03-stable
- d3schb2 appears in SCOPe 2.04
- d3schb2 appears in the current release, SCOPe 2.08

 View in 3D
View in 3D