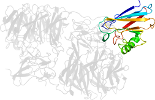
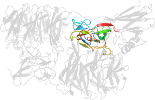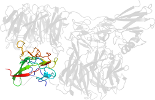Lineage for d2iaac_ (2iaa C:)
- Root: SCOPe 2.07

Class b: All beta proteins [48724] (178 folds) 
Fold b.6: Cupredoxin-like [49502] (2 superfamilies)
sandwich; 7 strands in 2 sheets, greek-key
variations: some members have additional 1-2 strands
Superfamily b.6.1: Cupredoxins [49503] (8 families) 
contains copper-binding site
Family b.6.1.1: Plastocyanin/azurin-like [49504] (10 proteins)
mono-domain proteins
Protein automated matches [190545] (9 species)
not a true protein
Species Alcaligenes faecalis [TaxId:511] [187784] (4 PDB entries) 
Domain d2iaac_: 2iaa C: [165454]
Other proteins in same PDB: d2iaab_, d2iaae_
automated match to d1dyza_
complexed with cu
Details for d2iaac_
PDB Entry: 2iaa (more details), 1.95 Å
PDB Description: Crystal Structure of an Electron Transfer Complex Between Aromatic Amine Dephydrogenase and Azurin from Alcaligenes Faecalis (Form 2)
PDB Compounds: (C:) AzurinSCOPe Domain Sequences for d2iaac_:
Sequence; same for both SEQRES and ATOM records: (download)
>d2iaac_ b.6.1.1 (C:) automated matches {Alcaligenes faecalis [TaxId: 511]}
acdvsiegndsmqfntksivvdktckeftinlkhtgklpkaamghnvvvskksdesavat
dgmkaglnndyvkagderviahtsvigggetdsvtfdvsklkegedyaffcsfpghwsim
kgtielgs
SCOPe Domain Coordinates for d2iaac_:
Click to download the PDB-style file with coordinates for d2iaac_.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d2iaac_:
- d2iaac_ first appeared in SCOPe 2.01
- d2iaac_ appears in SCOPe 2.06
- d2iaac_ appears in the current release, SCOPe 2.08

 View in 3D
View in 3D