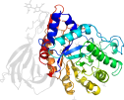
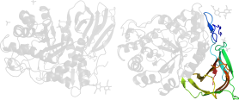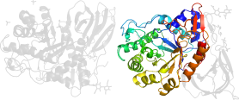Lineage for d1y7va1 (1y7v A:1-77,A:432-497)
- Root: SCOP 1.75

Class b: All beta proteins [48724] (174 folds) 
Fold b.71: Glycosyl hydrolase domain [51010] (1 superfamily)
folded sheet; greek-key
Superfamily b.71.1: Glycosyl hydrolase domain [51011] (5 families) 
this domain is C-terminal to the catalytic beta/alpha barrel domain
Family b.71.1.2: Composite domain of glycosyl hydrolase families 5, 30, 39 and 51 [89388] (4 proteins)
interrupted by the catalytic domain; the C-terminal core is similar to the alpha-amylase domain
Protein Glucosylceramidase [89389] (1 species)
glycosyl hydrolase family 30; the N-terminal extension wraps around the C-terminal core
Species Human (Homo sapiens) [TaxId:9606] [89390] (11 PDB entries) 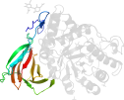
Domain d1y7va1: 1y7v A:1-77,A:432-497 [122725]
Other proteins in same PDB: d1y7va2, d1y7vb2
automatically matched to d1ogsa1
complexed with ins, nag, so4; mutant
Details for d1y7va1
PDB Entry: 1y7v (more details), 2.4 Å
PDB Description: x-ray structure of human acid-beta-glucosidase covalently bound to conduritol b epoxide
PDB Compounds: (A:) glucosylceramidaseSCOP Domain Sequences for d1y7va1:
Sequence; same for both SEQRES and ATOM records: (download)
>d1y7va1 b.71.1.2 (A:1-77,A:432-497) Glucosylceramidase {Human (Homo sapiens) [TaxId: 9606]}
arpcipksfgyssvvcvcnatycdsfdpptfpalgtfsryestrsgrrmelsmgpiqanh
tgtgllltlqpeqkfqkXqrvglvasqkndldavalmhpdgsavvvvlnrsskdvpltik
dpavgfletispgysihtylwhrq
SCOP Domain Coordinates for d1y7va1:
Click to download the PDB-style file with coordinates for d1y7va1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1y7va1:
- d1y7va1 first appeared in SCOP 1.73
- d1y7va1 appears in SCOPe 2.01
- d1y7va1 appears in the current release, SCOPe 2.08

 View in 3D
View in 3D