Lineage for d1spva_ (1spv A:)
- Root: SCOPe 2.07

Class c: Alpha and beta proteins (a/b) [51349] (148 folds) 
Fold c.50: Macro domain-like [52948] (1 superfamily)
3 layers: a/b/a; mixed beta-sheet of 6 strands, order 165243, strand 3 is antiparallel to the rest
Superfamily c.50.1: Macro domain-like [52949] (4 families) 

Family c.50.1.2: Macro domain [89724] (7 proteins)
found in different proteins, including macro-H2a histone and the Appr-1"-p processing enzyme
Protein Hypothetical protein YmbD [102530] (1 species) 
Species Escherichia coli [TaxId:562] [102531] (1 PDB entry) 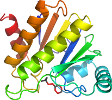
Domain d1spva_: 1spv A: [98957]
structural genomics; NESG target ER58
complexed with mes
Details for d1spva_
PDB Entry: 1spv (more details), 2 Å
PDB Description: crystal structure of the putative phosphatase of escherichia coli, northeast structural genomoics target er58
PDB Compounds: (A:) putative polyprotein/phosphataseSCOPe Domain Sequences for d1spva_:
Sequence; same for both SEQRES and ATOM records: (download)
>d1spva_ c.50.1.2 (A:) Hypothetical protein YmbD {Escherichia coli [TaxId: 562]}
trihvvqgditklavdvivnaanpslmggggvdgaihraagpalldaclkvrqqqgdcpt
ghavitlagdlpakavvhtvgpvwrggeqnedqllqdaylnslrlvaansytsvafpais
tgvygypraaaaeiavktvsefitrhalpeqvyfvcydeenahlyerlltqq
SCOPe Domain Coordinates for d1spva_:
Click to download the PDB-style file with coordinates for d1spva_.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1spva_:
- d1spva_ first appeared in SCOP 1.67
- d1spva_ appears in SCOPe 2.06
- d1spva_ appears in the current release, SCOPe 2.08
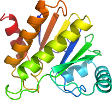
 View in 3D
View in 3D