Lineage for d1pj3a2 (1pj3 A:21-279)
- Root: SCOPe 2.02

Class c: Alpha and beta proteins (a/b) [51349] (147 folds) 
Fold c.58: Aminoacid dehydrogenase-like, N-terminal domain [53222] (1 superfamily)
core: 3 layers: a/b/a; parallel beta-sheet of 4 strands; 2134
Superfamily c.58.1: Aminoacid dehydrogenase-like, N-terminal domain [53223] (5 families) 

Family c.58.1.3: Malic enzyme N-domain [53240] (2 proteins)
Pfam PF00390; decorated with additional structures; includes N-terminal additional subdomains and extra N-terminal strand
Protein Mitochondrial NAD(P)-dependent malic enzyme [53241] (3 species) 
Species Human (Homo sapiens) [TaxId:9606] [53242] (10 PDB entries) 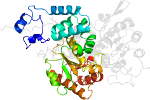
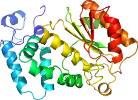
Domain d1pj3a2: 1pj3 A:21-279 [94727]
Other proteins in same PDB: d1pj3a1, d1pj3b1, d1pj3c1, d1pj3d1
complexed with fum, mn, nad, pyr
Details for d1pj3a2
PDB Entry: 1pj3 (more details), 2.1 Å
PDB Description: Crystal structure of human mitochondrial NAD(P)+-dependent malic enzyme in a pentary complex with natural substrate pyruvate, cofactor NAD+, Mn++, and allosteric activator fumarate.
PDB Compounds: (A:) NAD-dependent malic enzyme, mitochondrialSCOPe Domain Sequences for d1pj3a2:
Sequence; same for both SEQRES and ATOM records: (download)
>d1pj3a2 c.58.1.3 (A:21-279) Mitochondrial NAD(P)-dependent malic enzyme {Human (Homo sapiens) [TaxId: 9606]}
ikekgkplmlnprtnkgmaftlqerqmlglqgllppkietqdiqalrfhrnlkkmtsple
kyiyimgiqerneklfyrilqddieslmpivytptvglacsqyghifrrpkglfisisdr
ghvrsivdnwpenhvkavvvtdgerilglgdlgvygmgipvgklclytacagirpdrclp
vcidvgtdniallkdpfymglyqkrdrtqqyddlidefmkaitdrygrntliqfedfgnh
nafrflrkyrekyctfndd
SCOPe Domain Coordinates for d1pj3a2:
Click to download the PDB-style file with coordinates for d1pj3a2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1pj3a2:
- d1pj3a2 first appeared in SCOP 1.67
- d1pj3a2 appears in SCOPe 2.01
- d1pj3a2 appears in SCOPe 2.03
- d1pj3a2 appears in the current release, SCOPe 2.08
 View in 3D
View in 3D