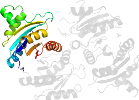
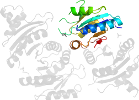
Lineage for d1jmva_ (1jmv A:)
- Root: SCOPe 2.07

Class c: Alpha and beta proteins (a/b) [51349] (148 folds) 
Fold c.26: Adenine nucleotide alpha hydrolase-like [52373] (3 superfamilies)
core: 3 layers, a/b/a ; parallel beta-sheet of 5 strands, order 32145
Superfamily c.26.2: Adenine nucleotide alpha hydrolases-like [52402] (7 families) 
share similar mode of ligand (Adenosine group) binding
can be subdivided into two group with closer relationships within each group than between the groups; the first three families form one group whereas the last two families form the other group
Family c.26.2.4: Universal stress protein-like [52436] (8 proteins)
Pfam PF00582
Protein Universal stress protein A, UspA [69461] (1 species) 
Species Haemophilus influenzae [TaxId:727] [69462] (1 PDB entry)
the nucleotide-binding is not known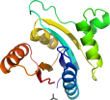
Domain d1jmva_: 1jmv A: [66897]
structural genomics
complexed with so4
Details for d1jmva_
PDB Entry: 1jmv (more details), 1.85 Å
PDB Description: Structure of Haemophylus influenzae Universal Stress Protein At 1.85A Resolution
PDB Compounds: (A:) Universal Stress Protein ASCOPe Domain Sequences for d1jmva_:
Sequence; same for both SEQRES and ATOM records: (download)
>d1jmva_ c.26.2.4 (A:) Universal stress protein A, UspA {Haemophilus influenzae [TaxId: 727]}
mykhilvavdlseespillkkavgiakrhdaklsiihvdvnfsdlytglidvnmssmqdr
istetqkalldlaesvdypiseklsgsgdlgqvlsdaieqydvdllvtghhqdfwsklms
strqvmntikidmlvvplrd
SCOPe Domain Coordinates for d1jmva_:
Click to download the PDB-style file with coordinates for d1jmva_.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1jmva_:
- d1jmva_ first appeared in SCOP 1.59
- d1jmva_ appears in SCOPe 2.06
- d1jmva_ appears in the current release, SCOPe 2.08

 View in 3D
View in 3D