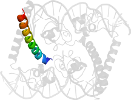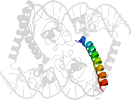Lineage for d1hwtg1 (1hwt G:59-97)
- Root: SCOPe 2.07
Class g: Small proteins [56992] (98 folds) 
Fold g.38: Zn2/Cys6 DNA-binding domain [57700] (1 superfamily)
all-alpha dimetal(zinc)-bound fold
Superfamily g.38.1: Zn2/Cys6 DNA-binding domain [57701] (1 family) 
duplication: two structural repeats are related by the pseudo dyad
Family g.38.1.1: Zn2/Cys6 DNA-binding domain [57702] (6 proteins) 
Protein Hap1 (Cyp1) [57709] (1 species) 
Species Baker's yeast (Saccharomyces cerevisiae) [TaxId:4932] [57710] (4 PDB entries) 
Domain d1hwtg1: 1hwt G:59-97 [45087]
Other proteins in same PDB: d1hwtc2, d1hwtd2, d1hwtg2, d1hwth2
protein/DNA complex; complexed with zn
Details for d1hwtg1
PDB Entry: 1hwt (more details), 2.5 Å
PDB Description: structure of a hap1/dna complex reveals dramatically asymmetric dna binding by a homodimeric protein
PDB Compounds: (G:) protein (heme activator protein)SCOPe Domain Sequences for d1hwtg1:
Sequence; same for both SEQRES and ATOM records: (download)
>d1hwtg1 g.38.1.1 (G:59-97) Hap1 (Cyp1) {Baker's yeast (Saccharomyces cerevisiae) [TaxId: 4932]}
riplscticrkrkvkcdklrphcqqctktgvahlchyme
SCOPe Domain Coordinates for d1hwtg1:
Click to download the PDB-style file with coordinates for d1hwtg1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1hwtg1:

 View in 3D
View in 3D