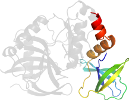
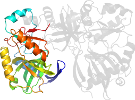
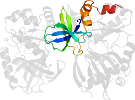Lineage for d1ckma2 (1ckm A:11-238)
- Root: SCOPe 2.07

Class d: Alpha and beta proteins (a+b) [53931] (388 folds) 
Fold d.142: ATP-grasp [56058] (2 superfamilies)
Consists of two subdomains with different alpha+beta folds
shares functional and structural similarities with the PIPK and protein kinase superfamilies
Superfamily d.142.2: DNA ligase/mRNA capping enzyme, catalytic domain [56091] (6 families) 
has a circularly permuted topology
Family d.142.2.3: mRNA capping enzyme [56100] (2 proteins)
automatically mapped to Pfam PF01331
Protein RNA guanylyltransferase (mRNA capping enzyme), N-terminal domain [56101] (1 species) 
Species Chlorella virus PBCV-1 [TaxId:10506] [56102] (3 PDB entries) 
Domain d1ckma2: 1ckm A:11-238 [41585]
Other proteins in same PDB: d1ckma1, d1ckmb1
protein/RNA complex; complexed with gtp
Details for d1ckma2
PDB Entry: 1ckm (more details), 2.5 Å
PDB Description: structure of two different conformations of mrna capping enzyme in complex with gtp
PDB Compounds: (A:) mRNA capping enzymeSCOPe Domain Sequences for d1ckma2:
Sequence; same for both SEQRES and ATOM records: (download)
>d1ckma2 d.142.2.3 (A:11-238) RNA guanylyltransferase (mRNA capping enzyme), N-terminal domain {Chlorella virus PBCV-1 [TaxId: 10506]}
nitteravltlnglqiklhkvvgesrddivakmkdlamddhkfprlpgpnpvsierkdfe
klkqnkyvvsektdgirfmmfftrvfgfkvctiidramtvyllpfkniprvlfqgsifdg
elcvdivekkfafvlfdavvvsgvtvsqmdlasrffamkrslkefknvpedpailrykew
iplehptiikdhlkkanaiyhtdgliimsvdepviygrnfnlfklkpg
SCOPe Domain Coordinates for d1ckma2:
Click to download the PDB-style file with coordinates for d1ckma2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1ckma2:

 View in 3D
View in 3D