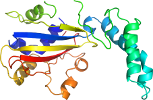
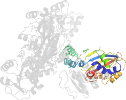
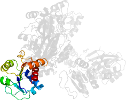
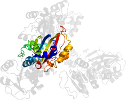
Lineage for d1brmc2 (1brm C:134-354)
- Root: SCOPe 2.08

Class d: Alpha and beta proteins (a+b) [53931] (396 folds) 
Fold d.81: FwdE/GAPDH domain-like [55346] (4 superfamilies)
core: alpha-beta-alpha-beta(3); mixed sheet: 2134, strand 2 is parallel to strand 1
Superfamily d.81.1: Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain [55347] (5 families) 
N-terminal domain is the classic Rossmann-fold
Family d.81.1.1: GAPDH-like [55348] (6 proteins)
has many additional secondary structures
Protein Aspartate beta-semialdehyde dehydrogenase [55361] (4 species) 
Species Escherichia coli [TaxId:562] [55362] (4 PDB entries)
Uniprot P00353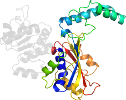
Domain d1brmc2: 1brm C:134-354 [39947]
Other proteins in same PDB: d1brma1, d1brmb1, d1brmc1
Details for d1brmc2
PDB Entry: 1brm (more details), 2.5 Å
PDB Description: aspartate beta-semialdehyde dehydrogenase from escherichia coli
PDB Compounds: (C:) aspartate-semialdehyde dehydrogenaseSCOPe Domain Sequences for d1brmc2:
Sequence, based on SEQRES records: (download)
>d1brmc2 d.81.1.1 (C:134-354) Aspartate beta-semialdehyde dehydrogenase {Escherichia coli [TaxId: 562]}
nctvslmlmslgglfandlvdwvsvatyqaasgggarhmrelltqmghlyghvadelatp
ssaildierkvttltrsgelpvdnfgvplagslipwidkqldngqsreewkgqaetnkil
ntssvipvdglcvrvgalrchsqaftiklkkdvsiptveellaahnpwakvvpndreitm
reltpaavtgtlttpvgrlrklnmgpeflsaftvgdqllwg
Sequence, based on observed residues (ATOM records): (download)
>d1brmc2 d.81.1.1 (C:134-354) Aspartate beta-semialdehyde dehydrogenase {Escherichia coli [TaxId: 562]}
nctvslmlmslgglfandlvdwvsvatyqaasgggarhmrelltqmghlyghvadelatp
ssaildierkvttltrsgelpvdnfgvplagslipwidkeewkgqaetnkilntssvipv
dglcvrvgalrchsqaftiklkkdvsiptveellaahnpwakvvpndreitmreltpaav
tgtlttpvgrlrklnmgpeflsaftvgdqllwg
SCOPe Domain Coordinates for d1brmc2:
Click to download the PDB-style file with coordinates for d1brmc2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1brmc2:
- d1brmc2 first appeared (with stable ids) in SCOP 1.55
- d1brmc2 appears in SCOPe 2.07
 View in 3D View in 3DDomains from other chains: (mouse over for more information) d1brma1, d1brma2, d1brmb1, d1brmb2 |