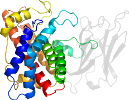
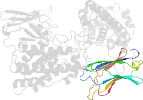
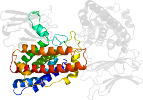
Lineage for d1qmdb2 (1qmd B:250-370)
- Root: SCOPe 2.06

Class b: All beta proteins [48724] (177 folds) 
Fold b.12: Lipase/lipooxygenase domain (PLAT/LH2 domain) [49722] (1 superfamily)
sandwich; 8 strands in 2 sheets; complex topology
duplication: has weak internal pseudo twofold symmetry
Superfamily b.12.1: Lipase/lipooxygenase domain (PLAT/LH2 domain) [49723] (4 families) 

Family b.12.1.3: Alpha-toxin, C-terminal domain [49738] (1 protein)
automatically mapped to Pfam PF01477
Protein Alpha-toxin, C-terminal domain [49739] (2 species) 
Species Clostridium perfringens, different strains [TaxId:1502] [49740] (6 PDB entries) 
Domain d1qmdb2: 1qmd B:250-370 [23650]
Other proteins in same PDB: d1qmda1, d1qmdb1
complexed with ca, zn
Details for d1qmdb2
PDB Entry: 1qmd (more details), 2.2 Å
PDB Description: calcium bound closed form alpha-toxin from clostridium perfringens
PDB Compounds: (B:) phospholipase cSCOPe Domain Sequences for d1qmdb2:
Sequence; same for both SEQRES and ATOM records: (download)
>d1qmdb2 b.12.1.3 (B:250-370) Alpha-toxin, C-terminal domain {Clostridium perfringens, different strains [TaxId: 1502]}
svgknvkelvayistsgekdagtddymyfgiktkdgktqewemdnpgndfmtgskdtytf
klkdenlkiddiqnmwirkrkytafsdaykpenikiiangkvvvdkdinewisgnstyni
k
SCOPe Domain Coordinates for d1qmdb2:
Click to download the PDB-style file with coordinates for d1qmdb2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1qmdb2:
- d1qmdb2 first appeared (with stable ids) in SCOP 1.55
- d1qmdb2 appears in SCOPe 2.05
- d1qmdb2 appears in SCOPe 2.07
- d1qmdb2 appears in the current release, SCOPe 2.08

 View in 3D
View in 3D