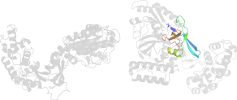
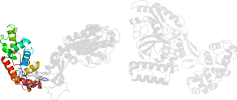Lineage for d1dgsa1 (1dgs A:401-581)
- Root: SCOPe 2.02

Class a: All alpha proteins [46456] (284 folds) 
Fold a.60: SAM domain-like [47768] (16 superfamilies)
4-5 helices; bundle of two orthogonally packed alpha-hairpins; involved in the interactions with DNA and proteins
Superfamily a.60.2: RuvA domain 2-like [47781] (7 families) 
duplication: contains two helix-hairpin-helix (HhH) motifs
Family a.60.2.2: NAD+-dependent DNA ligase, domain 3 [47786] (1 protein) 
Protein NAD+-dependent DNA ligase, domain 3 [47787] (1 species)
duplication: consists of two RuvA-like domains (four HhH motifs); also contains a zinc-finger subdomain
Species Thermus filiformis [TaxId:276] [47788] (2 PDB entries) 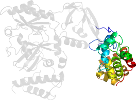
Domain d1dgsa1: 1dgs A:401-581 [17963]
Other proteins in same PDB: d1dgsa2, d1dgsa3, d1dgsb2, d1dgsb3
protein/DNA complex; complexed with amp, zn
Details for d1dgsa1
PDB Entry: 1dgs (more details), 2.9 Å
PDB Description: crystal structure of nad+-dependent dna ligase from t. filiformis
PDB Compounds: (A:) DNA ligaseSCOPe Domain Sequences for d1dgsa1:
Sequence; same for both SEQRES and ATOM records: (download)
>d1dgsa1 a.60.2.2 (A:401-581) NAD+-dependent DNA ligase, domain 3 {Thermus filiformis [TaxId: 276]}
rwpeacpecghrlvkegkvhrcpnplcpakrfeairhyasrkamdieglgeklierllek
glvrdvadlyhlrkedllglermgeksaqnllrqieeskhrglerllyalglpgvgevla
rnlarrfgtmdrlleasleelieveevgeltarailetlkdpafrdlvrrlkeagvsmes
k
SCOPe Domain Coordinates for d1dgsa1:
Click to download the PDB-style file with coordinates for d1dgsa1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1dgsa1:
- d1dgsa1 first appeared (with stable ids) in SCOP 1.55
- d1dgsa1 appears in SCOPe 2.01
- d1dgsa1 appears in SCOPe 2.03
- d1dgsa1 appears in the current release, SCOPe 2.08

 View in 3D
View in 3D