Lineage for d3kbma_ (3kbm A:)
- Root: SCOPe 2.01

Class c: Alpha and beta proteins (a/b) [51349] (147 folds) 
Fold c.1: TIM beta/alpha-barrel [51350] (33 superfamilies)
contains parallel beta-sheet barrel, closed; n=8, S=8; strand order 12345678
the first seven superfamilies have similar phosphate-binding sites
Superfamily c.1.15: Xylose isomerase-like [51658] (8 families) 
different families share similar but non-identical metal-binding sites
Family c.1.15.3: Xylose isomerase [51665] (2 proteins) 
Protein D-xylose isomerase [51666] (12 species) 
Species Streptomyces rubiginosus [TaxId:1929] [51670] (28 PDB entries) 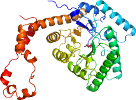
Domain d3kbma_: 3kbm A: [179238]
automated match to d1dxia_
complexed with cd, glc
Details for d3kbma_
PDB Entry: 3kbm (more details), 2 Å
PDB Description: room temperature x-ray structure of d-xylose isomerase complexed with 2cd(2+) co-factors and d12-d-alpha-glucose in the cyclic form
PDB Compounds: (A:) xylose isomeraseSCOPe Domain Sequences for d3kbma_:
Sequence; same for both SEQRES and ATOM records: (download)
>d3kbma_ c.1.15.3 (A:) D-xylose isomerase {Streptomyces rubiginosus [TaxId: 1929]}
mnyqptpedrftfglwtvgwqgrdpfgdatrraldpvesvrrlaelgahgvtfhdddlip
fgssdsereehvkrfrqalddtgmkvpmattnlfthpvfkdggftandrdvrryalrkti
rnidlavelgaetyvawggregaesggakdvrdaldrmkeafdllgeyvtsqgydirfai
epkpneprgdillptvghalafierlerpelygvnpevgheqmaglnfphgiaqalwagk
lfhidlngqngikydqdlrfgagdlraafwlvdllesagysgprhfdfkpprtedfdgvw
asaagcmrnylilkeraaafradpevqealrasrldelarptaadglqallddrsafeef
dvdaaaargmaferldqlamdhllgarg
SCOPe Domain Coordinates for d3kbma_:
Click to download the PDB-style file with coordinates for d3kbma_.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d3kbma_:
- d3kbma_ is new in SCOPe 2.01-stable
- d3kbma_ appears in SCOPe 2.02
- d3kbma_ appears in the current release, SCOPe 2.08
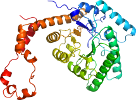
 View in 3D
View in 3D