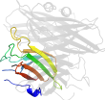
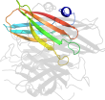
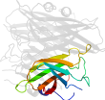
Lineage for d2hvbd_ (2hvb D:)
- Root: SCOPe 2.08

Class b: All beta proteins [48724] (180 folds) 
Fold b.1: Immunoglobulin-like beta-sandwich [48725] (33 superfamilies)
sandwich; 7 strands in 2 sheets; greek-key
some members of the fold have additional strands
Superfamily b.1.13: Superoxide reductase-like [49367] (2 families) 

Family b.1.13.1: Superoxide reductase-like [49368] (3 proteins)
binds iron in the site that is topologically equivalent to the copper-binding site of cupredoxins
automatically mapped to Pfam PF01880
Protein automated matches [190694] (3 species)
not a true protein
Species Pyrococcus horikoshii OT3 [TaxId:70601] [187828] (1 PDB entry) 
Domain d2hvbd_: 2hvb D: [165305]
automated match to d1do6a_
complexed with fe
Details for d2hvbd_
PDB Entry: 2hvb (more details), 2.5 Å
PDB Description: Crystal structure of hypothetical protein PH1083 from Pyrococcus horikoshii OT3
PDB Compounds: (D:) superoxide reductaseSCOPe Domain Sequences for d2hvbd_:
Sequence; same for both SEQRES and ATOM records: (download)
>d2hvbd_ b.1.13.1 (D:) automated matches {Pyrococcus horikoshii OT3 [TaxId: 70601]}
mlketirsgdwkgekhvpvieyeregdlvkvevsvgkeiphpntpehhiawielyfhpeg
gqfpilvgrveftnhsdpltepravfffktskkgklyalsycnihglwenevqle
SCOPe Domain Coordinates for d2hvbd_:
Click to download the PDB-style file with coordinates for d2hvbd_.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d2hvbd_:
- d2hvbd_ first appeared in SCOPe 2.01
- d2hvbd_ appears in SCOPe 2.07

 View in 3D
View in 3D