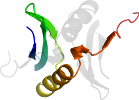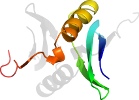Lineage for d2k7ia1 (2k7i A:1-62)
- Root: SCOPe 2.08

Class d: Alpha and beta proteins (a+b) [53931] (396 folds) 
Fold d.348: YegP-like [160112] (1 superfamily)
comprises two subunits or tandem repeats of beta(3)-alpha-beta motifs; these assemble with the formation of a single beta-sheet and swapping of the C-terminal strands
Superfamily d.348.1: YegP-like [160113] (2 families) 

Family d.348.1.1: YegP-like [160114] (4 proteins)
Pfam PF07411; DUF1508
Protein Uncharacterized protein Atu0232 [160115] (1 species)
homodimer
Species Agrobacterium tumefaciens [TaxId:358] [160116] (1 PDB entry)
Uniprot Q8UIR1 1-62
Domain d2k7ia1: 2k7i A:1-62 [148278]
Details for d2k7ia1
PDB Entry: 2k7i (more details)
PDB Description: solution nmr structure of protein atu0232 from agrobacterium tumefaciens. northeast structural genomics consortium (nesg) target att3. ontario center for structural proteomics target atc0223.
PDB Compounds: (A:) UPF0339 protein Atu0232SCOPe Domain Sequences for d2k7ia1:
Sequence; same for both SEQRES and ATOM records: (download)
>d2k7ia1 d.348.1.1 (A:1-62) Uncharacterized protein Atu0232 {Agrobacterium tumefaciens [TaxId: 358]}
mykfeiyqdkageyrfrfkasngetmfssegykakasaihaiesikrnsagadtvdlttm
ta
SCOPe Domain Coordinates for d2k7ia1:
Click to download the PDB-style file with coordinates for d2k7ia1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d2k7ia1:
- d2k7ia1 first appeared in SCOP 1.75
- d2k7ia1 appears in SCOPe 2.07
 View in 3D
View in 3D