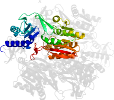Lineage for d1wxxd2 (1wxx D:65-382)
- Root: SCOP 1.73

Class c: Alpha and beta proteins (a/b) [51349] (141 folds) 
Fold c.66: S-adenosyl-L-methionine-dependent methyltransferases [53334] (1 superfamily)
core: 3 layers, a/b/a; mixed beta-sheet of 7 strands, order 3214576; strand 7 is antiparallel to the rest
Superfamily c.66.1: S-adenosyl-L-methionine-dependent methyltransferases [53335] (52 families) 

Family c.66.1.51: hypothetical RNA methyltransferase [142635] (4 proteins)
C-terminal part of Pfam PF03602; similar to (Uracil-5-)-methyltransferase (Pfam 05958)
Protein Hypothetical protein TTHA1280, middle and C-terminal domains [142638] (1 species) 
Species Thermus thermophilus [TaxId:274] [142639] (3 PDB entries) 
Domain d1wxxd2: 1wxx D:65-382 [121425]
Other proteins in same PDB: d1wxxa1, d1wxxb1, d1wxxc1, d1wxxd1
automatically matched to 1WXW A:65-382
complexed with k, po4
Details for d1wxxd2
PDB Entry: 1wxx (more details), 1.8 Å
PDB Description: Crystal structure of Tt1595, a putative SAM-dependent methyltransferase from Thermus thermophillus HB8
PDB Compounds: (D:) hypothetical protein TTHA1280SCOP Domain Sequences for d1wxxd2:
Sequence; same for both SEQRES and ATOM records: (download)
>d1wxxd2 c.66.1.51 (D:65-382) Hypothetical protein TTHA1280, middle and C-terminal domains {Thermus thermophilus [TaxId: 274]}
aedpvaallenlaqalarreavlrqdpeggyrlvhaegdllpglvvdyyaghavvqatah
awegllpqvaealrphvqsvlakndartreleglplyvrpllgevpervqvqegrvrylv
dlragqktgayldqrenrlymerfrgeraldvfsyaggfalhlalgfrevvavdssaeal
rraeenarlnglgnvrvleanafdllrrlekegerfdlvvldppafakgkkdverayray
kevnlraikllkeggilatascshhmteplfyamvaeaaqdahrllrvvekrgqpfdhpv
llnhpethylkfavfqvl
SCOP Domain Coordinates for d1wxxd2:
Click to download the PDB-style file with coordinates for d1wxxd2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1wxxd2:
- d1wxxd2 is new in SCOP 1.73
- d1wxxd2 appears in SCOP 1.75
- d1wxxd2 appears in the current release, SCOPe 2.08
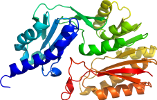
 View in 3D
View in 3D