Lineage for d1xmba2 (1xmb A:216-334)
- Root: SCOPe 2.05

Class d: Alpha and beta proteins (a+b) [53931] (381 folds) 
Fold d.58: Ferredoxin-like [54861] (59 superfamilies)
alpha+beta sandwich with antiparallel beta-sheet; (beta-alpha-beta)x2
Superfamily d.58.19: Bacterial exopeptidase dimerisation domain [55031] (1 family) 

Family d.58.19.1: Bacterial exopeptidase dimerisation domain [55032] (8 proteins) 
Protein IAA-amino acid hydrolase [117979] (1 species)
apparently monomeric
Species Thale cress (Arabidopsis thaliana) [TaxId:3702] [117980] (2 PDB entries)
Uniprot P54970 37-428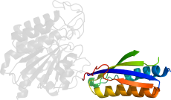
Domain d1xmba2: 1xmb A:216-334 [115480]
Other proteins in same PDB: d1xmba1
structural genomics target
Details for d1xmba2
PDB Entry: 1xmb (more details), 2 Å
PDB Description: x-ray structure of iaa-aminoacid hydrolase from arabidopsis thaliana gene at5g56660
PDB Compounds: (A:) IAA-amino acid hydrolase homolog 2SCOPe Domain Sequences for d1xmba2:
Sequence, based on SEQRES records: (download)
>d1xmba2 d.58.19.1 (A:216-334) IAA-amino acid hydrolase {Thale cress (Arabidopsis thaliana) [TaxId: 3702]}
agagvfeavitgkgghaaipqhtidpvvaassivlslqqlvsretdpldskvvtvskvng
gnafnvipdsitiggtlraftgftqlqqrvkevitkqaavhrcnasvnltpngrepmpp
Sequence, based on observed residues (ATOM records): (download)
>d1xmba2 d.58.19.1 (A:216-334) IAA-amino acid hydrolase {Thale cress (Arabidopsis thaliana) [TaxId: 3702]}
agagvfeavitgktidpvvaassivlslqqlvsretdpldskvvtvskvnpdsitiggtl
raftgftqlqqrvkevitkqaavhrcnasvnltpngrepmpp
SCOPe Domain Coordinates for d1xmba2:
Click to download the PDB-style file with coordinates for d1xmba2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1xmba2:
- d1xmba2 first appeared in SCOP 1.71
- d1xmba2 appears in SCOPe 2.04
- d1xmba2 appears in SCOPe 2.06
- d1xmba2 appears in the current release, SCOPe 2.08

 View in 3D
View in 3D